Abstract
The rock ptarmigan Lagopus muta is a grouse widely distributed in the arctic and subarctic regions of Eurasia and North America. The range of Japanese endemic subspecies (Japanese rock ptarmigans: L. m. japonica) is the southernmost of this species, but their origin is little known. Here, we determined the complete mitogenome of this subspecies and carried out the phylogenetic analysis. It was demonstrated that Japanese rock ptarmigans are old divergent lineage.
Keywords:
The rock ptarmigan (Lagopus muta) is a medium-sized grouse widely distributed in the arctic and subarctic regions of Eurasia and North America (del Hoyo et al. Citation1994). The Japanese rock ptarmigans (L. m. japonica) is an endemic subspecies of Japan and their habitat is limited to the high mountain area of middle Honshu island, where is the southernmost range of this species (Baba et al. Citation2001). Although the phylogeographic studies of this species were carried out (Holder et al. Citation2000; Caizergues et al. Citation2003; Lagerholm et al. Citation2017), the origin of Japanese rock ptarmigans is little known. Here, we determined the complete mitogenome of Japanese rock ptarmigans and carried out the phylogenetic analysis among the Lagopus muta and its related species.
The whole blood sample of the male Japanese rock ptarmigans was provided from Omachi Alpine Museum. The sample (RPT1) was stored in the Graduate School of Integrated Sciences for Life, Hiroshima University, Hiroshima, Japan. We determined the mitogenome by the procedures described in Nishibori et al. (Citation2001). The mitogenome of Japanese rock ptarmigans is 16,672 bp in length, and it has been deposited in DDBJ (LC528834). This sequence was aligned together with two mitogenomes of rock ptarmigans retrieved from NCBI and phylogenetic tree () was inferred by MEGA 7 program (Kumar et al. Citation2016) with the maximum-likelihood (ML) method.
Figure 1. The ML tree of rock ptarmigans based on mitogenome data. Bonasa bonasia and Lagopus lagopus were used as outgroup. The best nucleotide substitution model (GTR+I+Γ model) selected by the Bayesian Information Criterion was used for the tree inference. Accession numbers and scientific name of each sequences were shown. As for rock ptarmigans, the geographic locations were also shown in parentheses. The nodal numbers indicate the bootstrap probabilities with 1,000 replications.
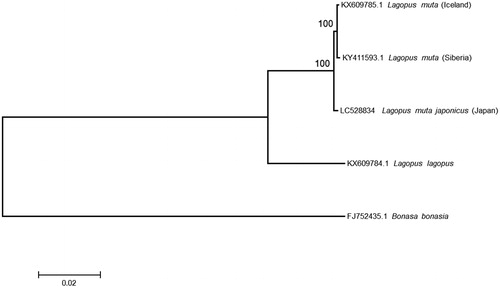
The rock ptarmigans from Iceland and Siberia formed a clade with 100% bootstrap probability and Japanese rock ptarmigans were placed as sister lineage of this clade. Lagerholm et al. (Citation2017) suggested Siberian, Arctic insular (Greenland, Svalbard, and Iceland) and North American populations are genetically close each other and form a distinct group from European population. Although only three rock ptarmigans (Siberia, Iceland, and Japan) were used in this study, our result suggests Japanese rock ptarmigans are old lineage before the rock ptarmigans spread to whole Siberia, Arctic islands, and North America.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Baba Y, Fujimaki Y, Yoshu R, Koike H. 2001. Genetic variability in the mitochondrial control region of the Japanese Rock Ptarmigan Lagopus mutus japonicus. Jpn J Ornithol. 50(2):53–64.
- Caizergues A, Bernard-Laurent A, Brenot JF, Ellison L, Rasplus JY. 2003. Population genetic structure of rock ptarmigan Lagopus mutus in Northern and Western Europe. Mol Ecol. 12(8):2267–2274.
- del Hoyo J, Elliott A, Sargatal J. 1994. Handbook of the birds of the world. Volume 2: new world vultures to guineafowl. Barcelona: Lynx Edicions.
- Holder K, Montgomerie R, Friesen VL. 2000. Glacial vicariance and historical biogeography of rock ptarmigan (Lagopus mutus) in the Bering region. Mol Ecol. 9(9):1265–1278.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lagerholm VK, Sandoval-Castellanos E, Vaniscotte A, Potapova OR, Tomek T, Bochenski ZM, Shepherd P, Barton N, Van Dyck MC, Miller R, et al. 2017. Range shifts or extinction? Ancient DNA and distribution modelling reveal past and future responses to climate warming in cold-adapted birds. Glob Chang Biol. 23(4):1425–1435.
- Nishibori M, Hayashi T, Tsudzuki M, Yamamoto Y, Yasue H. 2001. Complete sequence of the Japanese quail (Coturnix japonica) mitochondrial genome and its genetic relationship with related species. Anim Genet. 32(6):380–385.
