Abstract
In this research, on an Illumina platform, the full mitochondrial genome of Holothuria spinifera was listed in a sequence and also gathered by using the NovoPlasty v. 2.7.1. It was submitted to NCBI GenBank, and is available with accession number MN816440. The size of genome was 15,812 bp and contained 12 protein-coding genes, two rRNA genes, and 22 tRNA genes. The configuration of A + T in Holothuria spinifera mtDNA was 60.44%. Except five tRNAs and ND6, others are placed on the H-strand. By using the Neighbor-Joining method by software MEGA5.0, the phylogenetic relationship of 13 species of sea cucumber was analyzed. Holothuria spinifera was most closely associated to Parastichopus parvimensis.
Holothuria spinifera is a species of the family Holothuriidae. Holothuria spinifera being a benthic creature that feeds on debris and sediments species, it is living in the bottom of a thick sand or shell at a depth of 38–42 m (http://www.marinespecies.org/aphia.php?p=taxdetails&id=732394). It usually lives in Red Sea, Arabian Gulf, Sri Lanka, Philippines, Western Australia, and Southern China. By doing the comparison with other mitochondrial genomes of sea cucumber, it explains a scientific basis for the systematic evolution of sea cucumber. Therefore, it is very important for doing the complete research on mitochondrial genome of Holothuria spinifera.
The samples that were used for the sequencing were gathered in Sanya, Hainan Province, China (18°20′N 109°71′E) and kept at −80 °C before sequencing. We isolated the complete mitochondrial genome of Holothuria spinifera and made a list in the sequence on the basis of the Illumina pair-end technology, and it was assembled by using the SOAP denovo V2.04 software (http://soap.genomics.org.cn/). We forecasted gene by using the DOGMA software (http://dogma.ccbb.utexas.edu/). The genome contains rRNA and tRNA. MEGA5.0 (Kumar et al. Citation2016; Saitou and Nei Citation1987; Tamura et al. Citation2004) was put to use for many alignments. The Holothuria spinifera’s mtDNA map of the mitochondrial genome was drew by using the online tool OGDraw, which can be seen in https://chlorobox.mpimp-golm.mpg.de/OGDraw.html (Lohse et al. Citation2013). The specimen is stored in the Stock Enhancement in North China’s Sea and Key Laboratory of Mariculture, Ministry of Agriculture and Rural Affairs, Dalian Ocean University (voucher number: DLOU-KLM-SC02).
The size of the full Holothuria spinifera’s mitochondrial genome is 15,812 bp (GenBank registration number: MN816440), which includes two rRNA genes, 22 tRNA genes, and 12 protein-coding genes. The mitochondrial genome of Holothuria spinifera configuration is 27.26% for A, 15.73% for C, 23.84% for G, and 33.18% for T. The percentage of A + T in Holothuria spinifera mtDNA was 60.44%. Except ND6 and five tRNAs (tRNA-Ser, tRNA-Gln, tRNA-Ala, tRNA-Val, and tRNA-Asp), the others are placed on the H-strand. The initiation codon of most protein-coding genes was TTA (five of 12 genes) and CTA (four of 12 genes). Most termination codons of the protein are CAT (nine of 12 genes) or TAT (two of 12 genes), except for ND6. The 22 tRNA coding genes range in size from 63 bp to 70 bp.
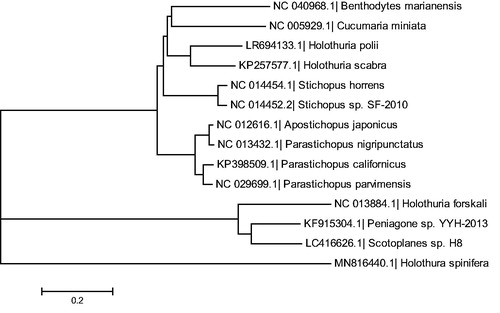
To ascertain their phylogenetic placements, we analyzed 13 species of sea cucumber using the Neighbor-Joining method by MEGA 7.0. The results revealed that Holothuria spinifera was closely related with Parastichopus parvimensis and the distance between Holothuria spinifera and Holothuria forskali was the farthest (). The mitochondrial genome of Holothuria spinifera provided valuable genomic, phylogeny, and taxonomy information of sea cucumber studies.
Acknowledgements
The authors thank for the materials provided by Researcher Chunhua Ren of South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kumar S, Stecher G, Tamura K. 2016. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4(4):406–425.
- Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 101(30):11030–11035.