Abstract
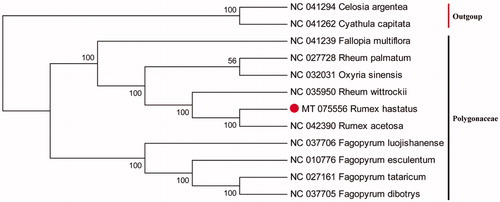
Rumex hastatus D. Don is a perennial shrub, which is distributed in Pakistan, Afghanistan, and the southwest of China. The complete chloroplast (cp) genome is 158,985 bp in length with an overall GC content of 37.4% containing a large single copy (LSC) region of 85,050 bp, a small single copy (SSC) region of 12,995 bp, and a pair of inverted repeats (IRs) region of 30,470 bp. Totally, 126 unique genes were predicted, including 81 protein-coding genes, 37 tRNA genes, and eight ribosomal RNA genes. Phylogenetic tree indicated that R. hastatus is closely related to R. acetosa.
Rumex hastatus D. Don is a perennial shrub, which belongs to the family of Polygonaceae. The plant is mainly distributed in Pakistan, Afghanistan, and the southwest of China, growing between 700 and 2500 m (Sahreen et al. Citation2011). As a traditional ethnic medicine, the whole plant is commonly used for treatment of lungs bleeding and skin disorder (Sahreen et al. Citation2014). In recent years, the research on R. hastatus was mainly focused on its chemical compositions, morphological taxonomy, and biological activities, etc., but there is no complete chloroplast (cp) genome of R. hastatus available in public databases. Hence, we first reported and characterized the cp genome of R. hastatus, which will provide useful genomic resource for further studies in conservation and evolution of R. hastatus.
Fresh leaves of R. hastatus were obtained from Cangshan Mountain (25°75′83″N, 100°11′73″E) of Dali Counties, Yunnan Province, China, and the voucher specimen was deposited in the herbarium of Dali University (LJ2020010504). Total DNA was extracted using the DNeasy plant mini kit (Qiagen, Hilden, Germany), and sequenced using Illumina NovaSeq system (San Diego, CA). Approximately, 8.4 Gb raw data (56,050,646 reads) were assembled by NOVOPlasty (Dierckxsens et al. Citation2017), and the assembled cp genome was annotated by GeSeq (Tillich et al. Citation2017). The annotated cp genome was submitted into GenBank with the accession number MT075556.
The cp genome of R. hastatus was 158,985 bp in length with an overall GC content of 37.4%, which contains a large single copy (LSC) region of 85,050 bp, a small single copy (SSC) region of 12,995 bp, and a pair of inverted repeats (IRs) regions of 30,470 bp. The genome comprises of 126 genes, including 81 protein-coding genes, 37 tRNA genes, and eight ribosomal RNA genes. To confirm the phylogenetic position of R. hastatus, the other 12 species of Polygonaceae subfamily from NCBI were aligned using MAFFT v7.307 (Katoh and Standley Citation2013). The maximum-likelihood (ML) tree was performed with RAxML v8.2.12 (Tan et al. Citation2019) using 1000 bootstraps. Celosia dargemtea and Cyathula capitata were served as the out-group. The phylogenetic tree showed that R. hastatus is closely related to R. acetosa (). This newly reported cp genome will be helpful for further phylogenetic study and evolutionary of Polygonaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 4(45):e18–e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Sahreen S, Khan MR, Khan RA. 2011. Phenolic compounds and antioxidant activities of Rumex hastatus D. Don. Leaves. J Med Plants Res. 13(5):2755–2765.
- Sahreen S, Khan MR, Khan RA. 2014. Comprehensive assessment of phenolics and antiradical potential of Rumex hastatus D. Don. roots. BMC Complement Altern Med. 1(14):47.
- Tan W, Gao H, Yang T, Jiang W, Zhang H, Yu X, Tian X. 2019. The complete chloroplast genome of a medicinal resource plant (Rumex crispus). Mitochondrial DNA B. 4(2):2800–2801.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq– versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.