Abstract
We sequenced the complete mitogenome of a specimen of Cymbiola nobilis (Neogastropoda, Volutidae) from Phú Quốc Island in Vietnam. The mitogenome of this predatory sea snail is 16,314-bp long and comprises the typical set of genes encoding 13 proteins, 2 rRNAs, and 22 tRNAs. It features a complex control region that contains two copies of an inverted repeat sequence separated by an AT-rich region, a characteristic previously observed in a few other gastropods. Phylogenetic analysis based on all protein-coding genes revealed that the Volutidae form a strongly supported clade.
The noble volute Cymbiola nobilis Lightfoot, 1786 (Neogastropoda, Volutidae) is a predatory sea snail that occurs throughout Asia. Adult specimens can reach up to 20 cm in length and feature a beige shell displaying brown stripes and a mantle with yellowish-black spots. As a consequence of human consumption, the seashell trade by volute collectors and also of shell utilization as material for crafting fine art, this marine gastropod is now considered to be a vulnerable species. In Vietnam, C. nobilis (Ốc Vôi in Vietnamese) is distributed from the southern part of the central coast to the southern extremity of the country (Nguyen et al. Citation1994). Here, we report the complete mitogenome sequence of a C. nobilis specimen that was collected on November 2018 from the Vietnamese Island of Phú Quốc (10°13′43ʺN; 103°57′27ʺE).
A piece of the specimen mantle was preserved in 95% ethanol and sent to the University of Szczecin (Poland) where DNA was extracted. The rest of the flesh and remaining shell are being kept at Saigon University (Vietnam) under the registration name SGU03. DNA sequencing was performed at the Beijing Genomics Institute (Shenzhen) on the BGISEQ-500 platform. Reads were assembled using SPAdes 3.12.0 (Bankevich et al. Citation2012), with a k-mer parameter of 85. A 15,288-bp mitogenome contig lacking the control region was initially identified and then extended to cover the entire mitogenome, using Consed (Gordon et al. Citation1998) and merged reads paired with FLASH (Magoc and Salzberg Citation2011).
The complete mitogenome of C. nobilis (GenBank MN871953) is 16,314-bp long and contains a total of 37 genes coding for 13 proteins, 2 rRNAs, and 22 tRNAs. All genes are oriented in the same direction and their order is identical to those of the other completely sequenced mitogenomes available for the Volutidae, Cymbium olla (Cunha et al. Citation2009), Melo melo (Zhong et al. Citation2019), and Neptuneopsis gilchristi (Harasewych et al. Citation2019). The control region of the C. nobilis mitogenome, located between cox3 and trnF, displays two copies of an inverted repeat sequence of about 500 bp that are separated by an AT-rich DNA segment. Long control regions with a similar structure have been observed in a few other Gastropoda mitogenomes where they account for their large size: for example, in the 16,887-bp mitogenome of Littorina saxatilis (Marques et al. Citation2017) and in the 16,430-bp genome of Conus quercinus (Gao et al. Citation2018).
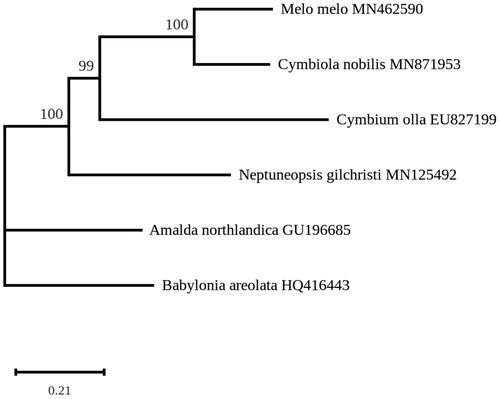
A phylogeny was inferred using the mitogenome sequences of 3 other taxa of the Volutidae, the Olividae Amalda northlandica and the Buccinidae Babylonia areolata as an outgroup. Concatenated protein-coding genes were aligned using MAFFT (Katoh and Standley Citation2016), and the resulting alignment was used for maximum likelihood analyses with RaxML 8.0 (Stamatakis Citation2014) with 1000 bootstrap replications. The best-scoring tree shows that C. nobilis and M. melo form a strongly-supported clade within a cluster containing all Volutidae, strongly separated from A. nortlandica and B. areolata ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Cunha RL, Grande C, Zardoya R. 2009. Neogastropod phylogenetic relationships based on entire mitochondrial genomes. BMC Evol Biol. 9(1):210.
- Gao B, Peng C, Chen Q, Zhang J, Shi Q. 2018. Mitochondrial genome sequencing of a vermivorous cone snail Conus quercinus supports the correlative analysis between phylogenetic relationships and dietary types of Conus species. PLOS One. 13(7):e0193053.
- Gordon D, Abajian C, Green P. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8(3):195–202.
- Harasewych MG, Sei M, Wirshing H, Gonzalez VL, Uribe J. 2019. The complete mitochondrial genome of Neptuneopsis gilchristi GB Sowerby III, 1898 (Neogastropoda: Volutidae: Calliotectinae). Nautilus. 133:67–73.
- Magoc T, Salzberg S. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27(21):2957–2963.
- Marques JP, Sotelo G, Larsson T, Johannesson K, Panova M, Faria R. 2017. Comparative mitogenomic analysis of three species of periwinkles: Littorina fabalis, L. obtusata and L. saxatilis. Mar Geonomics. 32:41–47.
- Nguyen HP, Ta MD, Pham TD, Dao TH, Vo ST, Bui TP, Tran TT. 1994. The main commercial sea products in the southern part of central Vietnam. Collection. V. Khánh Hòa – Vietnam. Nha Trang: Institute of Oceanography; p. 125–139.
- Katoh K, Standley DM. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics. 32(13):1933–1942.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zhong S, Huang G, Liu Y, Huang L. 2019. The complete mitochondrial genome of marine gastropod Melo melo (Neogastropoda: Volutoidea). Mitochondrial DNA Part B. 4(2):4161–4162.