Abstract
Artemisia ordosica is one of the important plants in succession of psammophytes in sandy (desert), it is a high ecological, economic and scientific research value. The chloproplast is photosynthetic organelles that provide energy for green plants. In this study, we reported the complete chloroplast (cp) genome sequence of A. ordosica, which were assembled using high-throughput sequencing technology. The results showed that the total cpDNA genome of A. ordosica is 151,209 bp in length, consisting of a large single copy (LSC) region of 82,980 bp, a small single copy (SSC) region of 18,303 bp, and a pair of inverted repeat regions (IRa and IRb, 24,963 bp each). Among all genes, the content of GC is 37.4%. The LSC, SSC, and IRs are 58.44%, 12.10%, and 33.02% of the A. ordosica cpDNA genome length respectively. Furthermore, the genome contained a total of 113 genes, comprising 79 protein-coding genes, 4 rRNA genes, and 29 tRNA genes. A phylogenetic analysis based on 21cpDNA genomes suggested that the A.ordosica is closely related to A. scoparia.
Introduction
Artemisia ordosica belongs to Artemisia of Compositae, which is an important and unique sand-fixing semi-shrub plant in China (Zhang et al. Citation2003). A. ordosica has rich nutritional value and high medicinal value. (Xiao et al. Citation2016). At present, many scientists have done a lot of research on the ecology and the determination of chemical components (Zhong et al. Citation2016; Hao et al. Citation2019). However, the characteristics of the complete chloroplast genome of A. ordosica has not been elucidated. Therefore, the fresh leaves of A. ordosica were used to analyze the cpDNA genome by the high-throughput sequencing technology in this study. We hope to provide some help for the study of A. ordosica in the future.
Materials and methods
Fresh A. ordosica leaves were collected in Gudimao Village, Qinhe Town, Yuyang District, Yulin City, Shaanxi Province, China (E 109°41′10.71″, N 38°18′19.61″; Height:1,080 m above sea level) in September 2019. The specimen of A. ordosica was saved at the Herbarium (Accession Number: 20190901Yl01) of College of Life Sciences, Yulin University, Shaanxi Province, China. Total DNA was extracted from the fresh leaves according to a modified cetyltrimethyl ammonium bromide (CTAB) method (Doyle and Doyle Citation1987). The subsequent high-throughput sequencing was completed with the IIIuminaHiSeq Ten system, with the Artemisia frigida (NC020607) cpDNA genome was used as the reference sequence to annotate. The A. ordosica cpDNA genome was annotated with Geneious program (Kearse et al. Citation2012) after its sequence was aligned with the reference cpDNA genome. The OGDRAW online was used to visualize the A. ordosica cpDNA genome (Lohse et al. Citation2013). The cpDNA genome sequences were aligned using MAFFT (Kazutaka et al. Citation2002). We used the MEGA 7.0 program (Kumar et al. Citation2016) to construct a phylogenetic tree with the cpDNA sequences from 20 other plants according to the Neighbour-joining (NJ) method, with a bootstrap value of 1000. The annotated A. ordosica cpDNA genome sequence has been deposited into the GenBank database (accession number: MN932370).
Results
The cpDNA genome of A. ordosica was 151,209 bp in length, similar with the chloroplast structure of most other plants, with a typical quadripartite structure (Shinozaki et al. Citation1986; Chao, et al. Citation2017). The genome contains a pair of inverted repeat regions (IRa and IRb, 24,963 bp each) separated by a large single copy region (LSC, 82,980 bp) and a small single copy region (SSC, 18,303 bp). Among all genes, the content of GC is 37.4%. The LSC, SSC and IR regions accounted for 54.88%, 12.10% and 33.02% of the cpDNA genome of A. ordosica, respectively. Furthermore, a total of 113 gene species were annotated for A. ordosica, including 79 protein-coding genes, 4 rRNA genes, and 29 tRNA genes.
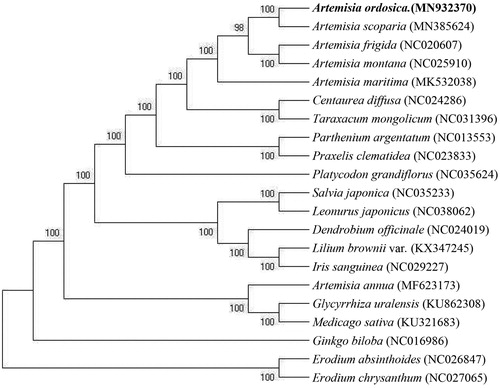
We constructed a phylogenetic tree based on 21 cpDNA genome sequences, with the Erodium absinthoides (NC026847) and Erodium chrysanthum (NC027065) cpDNA genome serving as an outgroup (). We aligned all 21 cpDNA genome sequences using MAFFT. The result showed that the A. ordosica (MN932370) is closely related to A. Scoparia (MN385624).
Figure 1. Phylogenetic tree constructed based on 21 species of chloroplast genome sequences. Accession numbers:Artemisia scoparia (MN385624); Artemisia frigida (NC020607); Artemisia montana (NC025910); Artemisia maritima (MK532038); Centaurea diffusa (NC024286); Taraxacum mongolicum (NC031396); Parthenium argentatum (NC013553); Praxelis clematidea (NC023833);Platycodon grandiflorus (NC035624); Salvia japonica (NC035233); Leonurus japonicas (NC038062); Dendrobium officinale (NC024019); Lilium brownii var. (KX347245);Iris sanguinea (NC029227);Artemisia annua (MF623173); Glycyrrhiza uralensis (KU862308); Medicago sativa (KU321683); Ginkgo biloba (NC016986); Erodium absinthoides (NC026847); Erodium chrysanthum (NC027065); Artemisia ordosica (MN932370).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chao X, Wenpan D, Wenqing L, et al. 2017. Comparative Analysis of Six Lagerstroemia Complete Chloroplast Genomes. Front Plant Sci. 8:15.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hao JS, Wang QH, Basenji RGL, Gong JH, Bao WQ, Bi LGT. 2019. Studies on the chemical constituents of Artemisia ordosica. China Phar Jou. 54(11):863–866.
- Kazutaka K, Kazuharu M, Kei-Ichi K, Takashi M. 2002. MAFFT:a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Aci Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. Mega 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW- a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucl Acids Res. 41(W1):W575–W581.
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. 1986. The complete nucleotide sequence of thetobacco chloroplast genome: its gene organization and expression. EMBO J. 5 (9):2043–2049.
- Xiao B, Bai JJ, Qi L, Lu LS, Tian XR, Yin J, Su YX. 2016. Research progress on the resource distribution, chemicalcomposition and pharmacological activity of Artemisia ordosica. China Pha. 2(13):1862–1864. Chinese.
- Zhang J, Ma JY, Yao J, Yang YL, Huang AL, Ji XH. 2003. Research on the resource utilization of Artemisia ordosica. China Wil Plan Res. 4(1):27–29.
- Zhong Y, Feng XS, Liu YR. 2016. Chemical constituents from Artemisia ordosica. Chem and Biol Eng. 33(3):36–38.
