Abstract
Medicago edgeworthii is a relatively unique species, which has the potential prospect to cross with M. sativa. In this study, we assembled the complete cp genome of M. edgeworthii using the Illumina sequencing data. The total genome is 122,442 bp in length and contains only one inverted repeat (IR) copy. The genome encodes 112 genes, including 76 protein-coding genes (PCGs), 32 tRNA genes, and 4 rRNA genes. Phylogenetic analysis using 11 cp genomes revealed the phylogenetic relationship of this species in Leguminosae.
Medicago edgeworthii, an allogamous diploid (2n = 2x = 16) perennials alfalfa, is indigenous to the Himalayas and alpine areas west to Afghanistan. Moreover, M. edgeworthii is a relatively unique species in Medicago, which is adapted to moist locations and extremely cold temperatures and has a remarkable ability to survive mechanical and physiological stress (Small and Jomphe Citation1989). So, it has the potential prospect to cross with M. sativa, which is the most widely cultivated forage throughout the world (Campbell and Bauchan Citation2002; Campbell Citation2003; Bouton Citation2007).
We sampled the leaf material from a plant specimen collected from Renbu county, Tibet (N 29.184433°, E89.911599°). And, now it is stored in the herbarium of Plant Biology Department, Beijing Forestry University with an accession number BFU-TB015. The whole genomic DNA was extracted by CTAB method (Doyle and Doyle Citation1987), and then the original data were obtained by next-generation sequencing technology. Clean reads were mapped to publish cp genomes of Medicagoas references in Geneious. Then, the filtered reads were de novo assembled into contigs. Finally, the complete chloroplast sequence was annotated using PGA (Qu et al. Citation2019). We have submitted it to the GenBank with the accession number MN901946.
The complete chloroplast genome of M. edgeworthii is 122,442 bp and only one copy of the IR region. There are 112 functional genes, including 76 protein-coding genes (PCGs), 32 tRNA genes, and 4 rRNA genes (rrn4.5, rrn5, rrn16, rrn23). Among them, 10 protein codes (rpl16, rpl2, ndhB, ndhA, ycf3, rpoC1, atpE, clpP, petB, petD) and 6 tRNA genes (trnI-GAU, tRNA-UGC, trnK-UUU, trnV-UAC, trnL-UAA, trnG-UUC) have introns. The base composition of the genome is 33.1% A, 16.4% C, 17.6% G, 33.0% T and the average GC content is 34.0%.
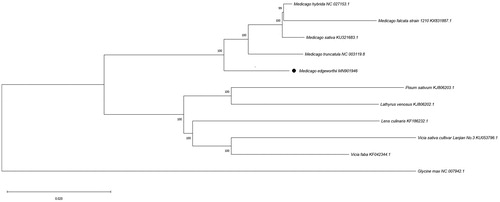
To further reveal the phylogenetic position of M. edgeworthiiin Leguminosae, we downloaded 11 complete chloroplast genome sequences of Leguminosae (Medicago hybrida, Medicago falcata strain 1210, Medicago truncatula, Medicago edgeworthii, Pisum sativum, Lathyrus venosus, Lens culinaris, Vicia sativa cultivar Lanjian No.3, Vicia faba, Glycine max) from GenBank and constructed a neighbour-joining (NJ) phylogenetic tree using MEGA X. We found M. edgeworthii clustered with other Medicago spp. with 100% bootstrap values ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bouton J. 2007. The economic benefits of forage improvement in the United States. Euphytica. 154(3):263–270.
- Campbell TA, Bauchan GR. 2002. Organelle based molecular analyses of the genetic relatedness of cultivated alfalfa (Medicago sativa L.) to Medicago edgeworthii Sirjaev, and Medicago ruthenica (L.) Ledebour. Euphytica. 125(1):51–58.
- Campbell TA. 2003. Investigation of variations in NBS motifs in alfalfa (Medicago sativa), M. edgeworthli, and M. ruthenica. Can J Plant Sci. 83(2):371–376.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Small E, Jomphe M. 1989. A synopsis of the genus Medicago (Leguminosae). Can J Bot. 67(11):3260–3294.