Abstract
We sequenced the complete mitogenome of the invasive flatworm Platydemus manokwari (Tricladida order, Geoplanidae family). This 19,959-bp genome contains 36 genes and is almost colinear with the mitogenomes of the two other species previously sampled from the Geoplanidae, Bipalium kewense and Obama nungara, both of which feature an extra tRNA gene. A single P. manokwari gene, encoding tRNA-Cys, is rearranged compared to its orthologs in the latter flatworms. Another distinctive trait of P. manokwari is the unusually large cox2 gene. In the phylogeny inferred from 12 proteins, P. manokwari formed a strongly supported clade with B. kewense and O. nungara.
Platydemus manokwari de Beauchamp, 1963 (Tricladida order, Geoplanidae family) is a large flatworm originating from New Guinea. It has been introduced both accidentally and willingly in many different areas, including the neighboring Pacific Islands, Asia, America and Europe (Justine et al. Citation2014, Citation2015). It is a known predator of snails, and when it was introduced on purpose to feed on invasive Gastropoda like the Giant African snail Lissachatina fulica, it also had a strong impact on endemic species (Sugiura Citation2009; Iwai et al. Citation2010; Sugiura and Yamaura Citation2010; Gerlach Citation2019). We sequenced the complete mitogenome of a specimen of P. manokwari collected in France and registered in the Muséum National d’Histoire Naturelle as MNHN_JL081, and from which a partial COI gene sequence was previously obtained (GenBank: KR349579).
A small portion of the specimen was sent to the Beijing Genomics Institute (BGI) in Shenzhen for DNA extraction and paired-end sequencing on the DNBSEQ platform. A total of 60 million 100-bp reads were generated; after filtering, they were assembled with SPAdes 3.12.0 (Bankevich et al. Citation2012) using a k-mer of 85. The contig corresponding to the mitogenome was retrieved, verified using the Consed package (Gordon et al. Citation1998), and annotated with the help of MITOS (Bernt et al. Citation2013). The rest of the specimen is available in the Muséum National d’Histoire Naturelle in Paris under the registration number MNHN_JL081.
The P. manokwari mitogenome (GenBank: MT081580) is 19,959-bp long and codes for 13 proteins, 2 rRNAs, and 21 tRNAs. It is almost perfectly colinear with the mitogenomes of the two other members of the Geoplanidae family for which complete mitogenomes are currently available, Bipalium kewense (Genbank: MK455837.1, Gastineau et al. Citation2019) and Obama nungara (sequence deposited in GenBank as Obama sp., Genbank: KP208777, Solà et al. Citation2015), which are invasive species too (Justine et al. Citation2018, Citation2020). The newly sequenced P. manokwari mitogenome displays three structural differences relative to its Geoplanidae counterparts. The gene coding for tRNA-Thr is missing and that coding for tRNA-Cys is rearranged, being located near cox3 instead of being adjacent to rrnS as in B. kewense and O. nungara, Moreover, at 1350 bp, the P. manokwari cox2 gene is twice as large compared to those of B. kewense (678 bp), O. nungara (780 bp) and flatworms belonging to closely related families. At the protein level, the extra nucleotides correspond to a region situated between a transmembrane domain and the copper A binding domain. As no detectable intron or intein was identified, this additional DNA sequence likely resulted from the expansion of a coding segment.
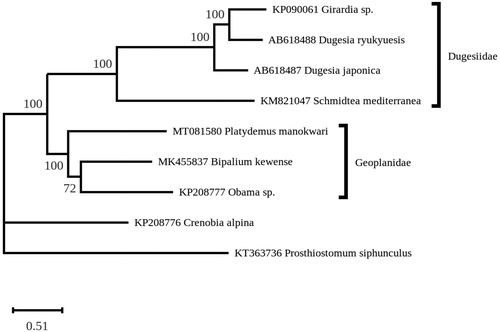
A maximum-likelihood phylogeny was inferred from 13 concatenated mitogenome-encoded proteins of eight platyhelminths from the Tricladida order using RAxML v.8.2.3 (Stamatakis Citation2014) and the MtArt substitution model (Abascal et al. Citation2007). A member of the Polycladida was used as outgroup. In the best tree, P. manokwari clustered strongly with B. kewense and Obama nungara, while all four members of the Dugesiidae family formed a highly supported sister clade (). This result is congruent with the revised classification of Sluys et al. (Citation2009), in which the Geoplanidae and Dugesiidae constitute the superfamily Geoplanoidea.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Abascal F, Posada D, Zardoya R. 2007. MtArt: a new model of amino acid replacement for Arthropoda. Mol Biol Evol. 24(1):1–5.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Gastineau R, Jean-Lou Justine J-L, Lemieux C, Monique Turmel M, Witkowski A. 2019. Complete mitogenome of the giant invasive hammerhead flatworm Bipalium kewense. Mitochondrial DNA Part B. 4(1):1343–1344.
- Gerlach J. 2019. Predation by invasive Platydemus manokwari flatworms: a laboratory study. Biol Lett. 54(1):47–60.
- Gordon D, Abajian C, Green P. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8(3):195–202.
- Iwai N, Sugiura S, Chiba S. 2010. Predation impacts of the invasive flatworm Platydemus manokwari on eggs and hatchlings of land snails. J Molluscan Stud. 76(3):275–278.
- Justine J-L, Winsor L, Barrière P, Fanai C, Gey D, Han AWK, La Quay-Velázquez G, Lee BPY, Lefevre J, Meyer J, et al. 2015. The invasive land planarian Platydemus manokwari (Platyhelminthes, Geoplanidae): records from six new localities. Including the First in the USA. Peer J. 3:e1037.
- Justine J-L, Winsor L, Gey D, Gros P, Thévenot J. 2014. The invasive New Guinea flatworm Platydemus manokwari in France, the first record for Europe: time for action is now. Peer J. 2:e297.
- Justine J-L, Winsor L, Gey D, Gros P, Thévenot J. 2018. Giant worms chez moi! Hammerhead flatworms (Platyhelminthes, Geoplanidae, Bipalium spp., Diversibipalium spp.) in metropolitan France and overseas French territories. Peer J. 6:e4672.
- Justine J-L, Winsor L, Gey D, Gros P, Thévenot J. 2020. Obama chez moi! The invasion of metropolitan France by the land planarian Obama nungara (Platyhelminthes, Geoplanidae). Peer J. 8:e8385.
- Sluys R, Kawakatsu M, Riutort M, Baguñà J. 2009. A new higher classification of planarian flatworms (Platyhelminthes, Tricladida). J Nat Hist. 43(29–30):1763–1777.
- Solà E, Álvarez-Presas M, Frías-López C, Littlewood DT, Rozas J, Riutort M. 2015. Evolutionary analysis of mitogenomes from parasitic and free-living flatworms. PLoS One. 2010(3):e0120081.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sugiura S. 2009. Seasonal fluctuation of invasive flatworm predation pressure on land snails: Implications for the range expansion and impacts of invasive species. Biol Conserv. 142:3013–3019.
- Sugiura S, Yamaura Y. 2010. Potential impacts of the invasive flatworm Platydemus manokwari on arboreal snails. In: Kawakami K., Okochi I, editors. Restoring the Oceanic Island ecosystem. Tokyo (Japan): Springer.