Abstract
Sauropus spatulifolius Beille is one kind of Chinese herbal medicine with anti-inflammatory and analgesic activities. In this study, we reported the complete chloroplast genome of S. spatulifolius Beille, and assembled and annotated with high-throughput sequencing data, which would provide help for its taxonomy research. The chloroplast sequence was 154,707 bp , with two of 87,438 bp and 19,427 bp single-copy regions, which were separated by two inverted repeat regions with 23,921 bp . A total of 129 genes were predicted, with GC content of 36.61%. Phylogenetic analysis showed that S. spatulifolius Beille closest to Glochidion chodoense in Malpighiales.
Sauropus spatulifolius Beille is an important national medicinal and a local food material for soup and Guangdong Herbal Tea in south of China (Commission Citation2015), which is distributed in southeast Asia (Zou et al. Citation2013). Its main component is Kaempferol-3-o-gentian glycoside, which has anti-inflammatory and analgesic, anti-allergic, anti HBe Ag and other active effects. (Parveen et al. Citation2007). Genomic information is important for the identification and classification of Euphorbiaceae plants, but the genome of S. spatulifolius Beille was deficient. The complete chloroplast genome was be used to identify a series of genus-specific DNA barcode sequences in medical plants. In this study, we reported the complete chloroplast sequence of S. spatulifolius Beille, which would be useful for its detection and evolutionary research.
The leaves of S. spatulifolius Beille were collected from the Zhongshan Yongzheng Chinese Herbal Medicine Research Institute (22°31′20.1ʺN, 113°24′2.40ʺE), Zhongshan city, Guangdong province, China. The samples were frozen by liquid nitrogen and stored at −80 °C refrigerator until used in the key laboratory for crops genetic improvement of Guangdong. The genomic DNA from S. spatulifolius Beille leaves was extracted using plant genomic DNA kit (Omega) and sequenced on the Novaseq platform (Illumina, San Diego, CA) according to the method recommended by the manufacturer. The sequence was assembled by the GetOrganelle (Jin et al. Citation2019) and annotated with the Geseq (Tillich et al. Citation2017), then submitted to GenBank with the accession number MT089915.
The complete chloroplast genome of S. spatulifolius Beille obtained in this study was 154,707 bp with a large and a small single-copy regions of 87,438 bp and 19,427 bp, which were separated by the same inverted repeat (IR) region of 23,921 bp. Total of 129 genes were predicted, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The GC content was 36.99% of the genome. The ACG was the start codon of ndhD gene.
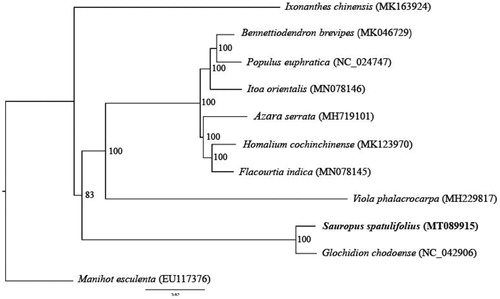
To compare the relationship between S. spatulifolius Beille and other 10 genera in the Malpighiales, a phylogenetic tree was constructed with the full chloroplast sequence of these species (Ixonanthes chinensis, Bennettiodendron brevipes, Populus euphratica, Itoa orientalis, Azara serrata, Homalium cochinchinense, Flacourtia indica, Viola phalacrocarpa, Glochidion chodoense and Manihot esculenta), which were obtained from NCBI GenBank, using MAFFT (Katoh and Standley Citation2013). Then, RAxML was used to construct a maximum likelihood tree (Stamatakis Citation2014) (). The results showed that S. spatulifolius Beille was closest to Glochidion chodoense.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Commission CP. 2015. Pharmacopoeia of the People’s Republic of China. (English version), Vol. I. Beijing, China: China Medical Science Press.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2019. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv. 256479
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Parveen Z, Deng Y, Saeed MK, Dai R, Ahamad W, Yu YH. 2007. Antiinflammatory and analgesic activities of Thesium chinense Turcz extracts and its major flavonoids, kaempferol and kaempferol-3-O-glucoside. Yakugaku Zasshi. 127(8):1275–1279.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zou YS, Foubert K, Tuenter E, Lemière F, Cos P, Maes L, Smits JM, Gelder R, Apers S, Pieters L. 2013. Antiplasmodial and cytotoxic activities of Striga asiatica and Sauropus spatulifolius extracts, and their isolated constituents. Phytochem Lett. 6(1):53–58.