Abstract
The complete chloroplast genome sequence of Bartramia pomiformis, a species of moss in the Bartramiaceae family, was determined using Illumina HiSeq paired-end sequencing data. The total size of the chloroplast genome of this species was 125,866 bp, and it contained a large single-copy (LSC) region of 87,214 bp, a small single-copy (SSC) region of 18,651 bp, and a pair of identical inverted repeat regions (IRs) of 10,014 bp. The genome contained 82 protein-coding genes, 36 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. The information of the B. pomiformis chloroplast genome will be valuable for evolutionary studies of the Bartramiaceae.
Bartramiaceae is a large family of moss that is considered the most conspicuous among its counterparts (Ochyra et al. Citation2008). The genus Bartramia was characterized by Hedwig (1801) and published in Species Muscorum Frondosorum. Bartramia pomiformis, or apple moss, is named after the round green capsules that emerge from its cushions (tufts) and make it highly distinguishable. This species prefers shady and acidic rocky areas such as rock outcrops or cracks but also grows on calcium enriched rocks (Fransen Citation2004). Apple moss is common, with both B. pomiformis and B. ithyphylla recognized in South Korea (Choe Citation1980); however, it has a wide-range distribution and slightly varies in the extent of its foliation and seta length.
Fresh samples were collected from Yeongyang-gun County (36° 38′ 19.04 N; 129° 9′ 24.82 E), Gyeongsangbuk-do Province, Korea and stored at Jeonbuk National University (JNU Herbarium) in Jeonju, Korea with the accession number YYJ 20191021-1. Genomic DNA was purified using a DNeasy® Plant Mini Kit (Qiagen, Hilden, Germany) and read by paired-end (PE) sequencing via the Illumina HiSeq platform (Illumina, San Diego, CA). De novo assembly and gene annotation were performed according to the methods described by Choi et al. (Citation2020). The phylogenetic tree was constructed based on analysis of maximum-likelihood estimates (ML) (JTT matrix-based model, MEGA7) using the concatenated alignment of 16 protein-coding genes from eight Bryophytes, two Marchantiophytes, and one Anthocerotophyte. Moreover, internal branches were assessed using 1000 bootstrap replicates of ML analyses.
The chloroplast (cp) genome of B. pomiformis (MT024676) has a total length of 15,584 bp, with 29.3% G + C content; the base composition is as follows: A (35.3%), C (14.7%), G (14.6%), and T (35. 5%). The cp genome of this organism consists of 87,214 bp large single-copy (LSC) and 18,651 bp small single-copy (SSC) regions separated by a pair of 10,014 bp inverted repeat (IR) regions. It also contains 82 protein-coding genes (PCGs), 36 tRNAs, and eight rRNAs, which are similar to conventional Bryophyta chloroplast genome values.
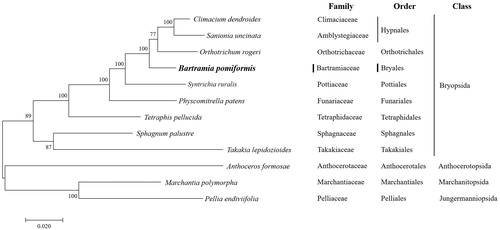
In conclusion, analysis of B. pomiformis datasets provides convincing evidence that supports many traditionally recognized families and identifies a higher level of phylogenetic structure () (Cox et al. Citation2000; Magombo Citation2003; Liu et al. Citation2014). Using our sequencing results and publicly available GenBank data, we confirmed the phylogenetic relationships among the members of the Bartramiaceae family belonging to the class Bryopsida. Therefore, whole chloroplast genome sequences provided sufficient genetic information for species identification and phylogenetic reconstruction of genus Bartramia.
Figure 1. Phylogenetic position of B. pomiformis determined by Maximum-likelihood methods based on combined analysis with amino acids sequences of 16 mitochondrial genes common in all taxa. The bootstrap values (1000) are presented near the corresponding branch. Branches that were supported by above 50% bootstrap values are indicated by bold lines. Sequences from 2 Marchantiopsida and 1 Anthocerotophyta were used as outgroup. GenBank accession numbers of mitogenomes used are Anthoceros formosae (NC_004543), Bartramia pomiformis (MT024676), Climactium dendroides (MT006132), Marchantia polymorpha (NC_037507), Orthotrichum rogeri (NC_026212), Pellia endiviifolia (NC_019628), Physcomitrella patens (NC_037465), Sphagnum palustre (NC_03019), Syntrichia filaris (MK852705), Takakia lepidozioides (NC_028738) and Tetraphis pellucida (NC_024291).
Disclosure statement
There are no conflicts of interest for all the authors including the implementation of research experiments and writing this article.
References
- Choe DM. 1980. Illustrated Flora and Fauna of Korea. Vol. 24. Seoul: Musci-Hepaticae., Ministry of Education; pp. 790.
- Choi Y, Han Y-D, Moon JC, Yoon Y-J. 2020. The complete mitochondrial genome of a moss Korea Climacium dendroides (Hedw.) F. Weber & D. Mohr. Mitochondrial DNA B. 5(1):1071–1072.
- Cox CJ, Goffinet B, Newton AE, Shaw AJ, Hedderson TA. 2000. Phylogenetic relationships among the diplolepideous-alternate mosses (Bryidae) inferred from nuclear and chloroplast DNA sequences. The Bryologist. 103(2):224–241.
- Fransen S. 2004. A taxanomic revision of Bartramia Hedw. Section Bartramia. Lindbergia. 29:113–122.
- Liu Y, Rafael M, Goffinet B. 2014. 350 Million years of mitochondrial genome stasis in mosses, and early land plant lineage. Mol Biol E. 31(10):2586–2591.
- Magombo ZL. 2003. The phylogeny of basal peristomate mosses: evidence from cpDNA, and implications for peristome evolution. Syst Bot. 28(1):24–38.
- Ochyra R, Lewis S, Bednarek-Ochyra H. 2008. The illustrated moss flora of Antarctica. Cambridge: Cambridge University Press; pp. 685.
