Abstract
The complete mitochondrial genome of Colochirus quadrangularis was obtained and described in this study. This complete mitochondrial genome is 17,157 bp in length and consists of 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes. Except ND6 and 5 tRNAs, the others were encoded by the heavy strand. The overall base composition of the heavy-strand was 40.23% A, 11.31% G, 21.76% C, and 26.71% T, with a high G + C content of 33.07%. The phylogenetic analysis suggested that C. quadrangularis was closest to Cucumaria miniata. The newly described mitochondrial genome may provide valuable data for the phylogenetic analysis for Holothuroidea.
Holothuroidea, known as sea cucumber, has gained a lot of attention from researchers worldwide because of their nutritive values as well as their beneficial influence on human health and possible therapeutic uses (Yuri Citation2018). Colochirus quadrangularis is a member of cucumaria. It is naturally distributed in the Indo-Pacific region, especially in the south of China, Sri Lanka, Indonesia, Australian, and so on (Liao Citation1997). Colochirus quadrangularis was found mostly in a variety of habitats from intertidal zone to deep seagrass bed (up to 100 m) (Liao Citation1997). Artificial breeding and larval rearing of C. quadrangularis was reported (Ajith Kumara et al. Citation2016).Unfortunately, only a DNA sequence COX1 of C. quadrangularis was reported earlier (O’Loughlin et al. Citation2016). Lack of genetic resources has hindered the conservation and utilization of C. quadrangularis. The genetic makers from mitochondrial DNA were very effective, which could successfully be applied to the study of population genetics, molecular phylogenetics, and evolutionary studies (Galtier et al. Citation2009). In this study, the complete mitochondrial genome of C. quadrangularis and its phylogenetic relationships within other sea cucumbers have been investigated.
The specimen was collected from Shenzhen, Guangdong province, China (N22°35′, E114°31′) and kept in the Zoological Herbarium of the Lingnan Normal University (Acc. Number SC20190819-1). The dorsal body muscle of C.quadrangularis were fixed in 100% ethanol and stored at-20 °C. Approximately 30 mg of muscle tissue was used for mitochondrial DNA (mtDNA) extraction with TIANamp Marine Animals DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s specification. MtDNA were sequenced using the Illumina Hiseq Sequencing System (Illumina Inc., San Diego, CA). The clean data were acquired and assembled by the SPAdes and PRICE (Bankevich et al. Citation2012). BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) and ORFs finder (https://www.ncbi.nlm.nih.gov/orffinder/) were used to identify and annotated protein-coding genes (PCGs). tRNAscan-SE 2.0 (Lowe and Chan Citation2016) was used to identified tRNA genes. A phylogenetic tree was constructed using MEGA 6.0 software.
The mitochondrial genome of C. quadrangularis is 17,157 bp in length (GenBank accession number: MT108721) and containing the typical set of 13 PCGs, 22 tRNA, and 2 rRNA genes, and two putative control regions. The heavy strand consists of 40.23% A, 11.31% G, 21.76% C, and 26.71% T bases, with a high A + T content of 66.93%. Of the 37 genes, 31 were encoded by the heavy strand and the others were encoded by the light strand. All PCGs were initiated by ATG. Eleven PCGs were terminate with the TAA as stop codon, while ND5and ND6 ended with TAG. Twenty-two tRNA genes ranged in size from 67 to 73 bp.
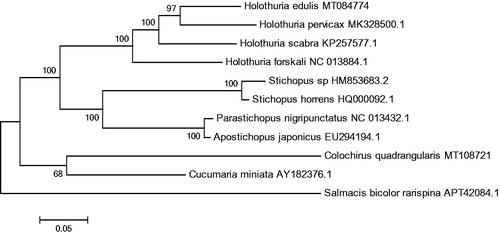
The phylogenetic tree was constructed based on 13 concatenated PCGs from 11 sea cucumbers from GenBank database, by maximum likelihood (ML) method. Salmacis bicolor rarispina was used as an outgroup for tree rooting (). It was demonstrated that C. quadrangularis was clustered with Cucumaria miniata, which revealed that C. quadrangularis was most affinitive to C. miniata. In conclusion, this genome will contribute to future phylogenetic studies and population genetic analyses for C. quadrangularis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ajith Kumara PAD, Jayanatha JS, Pushpakumara J, Bandara W, Dissanayake D. 2016. Artificial breeding and larval rearing of three tropical sea cucumber species – Holothuria scabra, Pseudocolochirus violaceus and Colochirus quadrangularis – in Sri Lanka. SPC Beche-de-Mer Infor Bul. 33:30–37.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Galtier N, Nabholz B, Glemin S, Hurst G. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol. 18(22):4541–4550.
- Liao Y. 1997. Fauna sincia: phylum Echinodermata class Holothuroidea. Beijing: Science Press.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: search and contextual analysis of transfer RNA Genes. Nucleic Acids Res. 44(W1):W54–57.
- O’Loughlin PM, Harding C, Paulay G. 2016. The sea cucumbers of Camden Sound in northwest Australia, including four new species (Echinodermata: Holothuroidea). MMV. 75:7–52.
- Yuri K. 2018. Pharmacological potential of sea cucumbers. Int J Mol Sci. 19(5):1342.