Abstract
A new super passion fruit, named ‘Ziyan,’ is an artificial hybrid of Passiflora. edulis Sims × P. edulis f. edulis Sims. In this paper, the complete chloroplast genome of the artificial hybrid ‘Ziyan’ was characterized using the Illumina sequencing technology. The entire chloroplast genome was 151,316 bp in length with 37% GC content and 131 genes were produced, including 86 protein-coding genes, 8 ribosomal RNA genes, and 37 transfer RNA genes. Phylogenetic analyses confirmed that ‘Ziyan’ was closely related to yellow passion flower ‘IAPAR-123’ and belongs to Passifloraceae, especially the genus Passiflora.
Passion fruit (Passiflora edulia Sims), called Baixiang-guo, belongs to the family Passifloraceae, and originated from tropical America. Passion fruit is a species of tropical climate and is distributed widely in Latin America, Brazil, Colombia, and China (Benincá et al. Citation2007). In the early twentieth century, Passiflora edulis Sims, as the new darling of the fruit market, was introduced to China, mainly in Taiwan, Guangdong, Guangxi, and Fujian (Rudnicki et al. Citation2007; Bernacci et al. Citation2008). In recent years, it has become one of the most popular fruits worldwide, with important economic significance due to its organoleptic and bioactive properties and high nutrient content. In the current market, yellow and purple passion fruits are both sold as the main varieties. The entire chloroplast (cp) genomes of yellow and purple passion fruits were reported by Cauz-Santos et al. (Citation2017) and Yang et al. (Citation2019). However, there is no complete chloroplast genome information for their hybrid. In this work, we have assembled the complete chloroplast (cp) genome of the artificial hybrid ‘Ziyan’, which provides useful insights into the genetic diversity and evolution of passion fruit.
Fresh leaves of ‘Ziyan’ were collected from the Fruit Research Institute, Fujian Academy of Agricultural Sciences, Fuzhou, China (Fujian, China, 119°19′57″E, 26°7′47″N), and were deposited at Nanjing Forestry University (No. NFUFG001). Total genomic DNA was extracted using the modified CTAB protocol (Yanqiong et al. Citation2019) and used to construct an Illumina paired-end library. In addition, the genomic DNA was sequenced using an Illumina HiSeq 2000 platform system (Illumina, San Diego, CA, USA) at Beijing Genomics Institute (BGI, Shenzhen, China), generating approximately 4.51 GB of raw data. After filtration, high-quality clean reads of 4.5 GB generated using the FastQC software (Andrews Citation2014) were used to assemble the cp genome using SPAdes version 3.9.0 (http://bioinf.spbau.ru/spades) (Bankevich et al. Citation2012) and the cp genome was annotated using the online program GeSeq (Tillich et al. Citation2017). The cp genome sequence of ‘Ziyan’ was deposited in GenBank under the accession number MT140635.
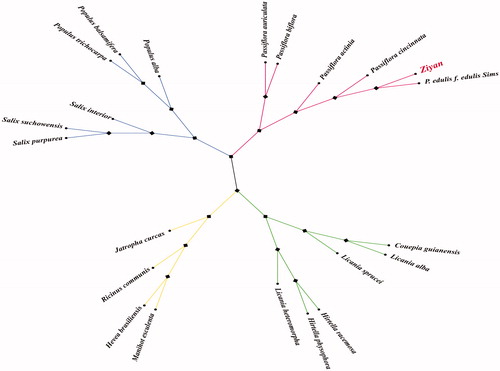
The circular genome of ‘Ziyan’ is 151,316 bp in size with a GC content of 37%. The genome structure exhibits a typical quadripartite feature comprising a large single-copy region (LSC) of 86,628 bp, a small single-copy region (SSC) of 13,378 bp, and a pair of inverted repeat regions (IR) of 26,155 bp. Totally, 131 unique genes were annotated in the cp genome, including 86 protein-coding genes, 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. In the IR regions, five genes (ycf1, rrn4.5, rrn5, rrn16, and rrn23) were found duplicated in each. Five genes of the annotated genes (atpF, ndhA, ndhB, rpl2, rpoC1) contained one intron, and 2 genes (clpP, ycf3) contained two introns. To further understand the evolution of the hybrid passion fruit ‘Ziyan’, the complete cp genomes of the 22 species were selected to construct phylogenic maximum-likelihood trees using RaxML software version 8.2.9. The bootstrap values were calculated using 1000 replicates (Stamatakis Citation2014). As shown in , phylogenetic analysis revealed that ‘Ziyan’ is clustered with the yellow passion flower ‘IAPAR-123’, within Passifloraceae, which is in agreement with a previous study (Yang et al. Citation2019).
Figure 1. Phylogenetic tree based on 22 complete cp genome sequences. Accession numbers: Populus alba (NC_008235.1), Populus trichocarpa (NC_009143.1), Populus balsamifera (NC_024735.1), Salix purpurea (NC_026722.1), Salix suchowensis (NC_026462.1), Salix interior (NC_024681.1), Manihot esculenta (NC_010433.1), Hevea brasiliensis (NC_015308.1), Ricinus communis (NC_016736.1), Jatropha curcas (NC_012224.1), Hirtella physophora (NC_024066.1), Hirtella racemosa (NC_024060.1), Licania heteromorpha (NC_024062.1), Licania alba (NC_024064.1), Licania sprucei (NC_024065.1), Couepia guianensis (NC_024063.1), Ziyan (MT_140635), P. edulis f. edulis Sims (KX290855.1), Passiflora cincinnata (KY820583.1), Passiflora actinia (MF807934.1), Passiflora auriculata (MF807936.1), Passiflora biflora (MF807937.1).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Andrews S. 2014. FastQC: a quality control tool for high throughput sequence data. [accessed 2020 Mar 27]. http://wwwbioinformaticsbabrahamacuk/projects/fastqc/.
- Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov A, Lesin V, Nikolenko S, Pham S, Prjibelski A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Benincá J, Montanher A, Zucolotto S, Schenkel E, Fröde T. 2007. Evaluation of the anti-inflammatory efficacy of Passiflora edulis. Food Chem. 104(3):1097–1105.
- Bernacci L, Soares-Scott M, Junqueira N, Passos I, Meletti L. 2008. Passiflora edulis Sims: the correct taxonomic way to cite the yellow passion fruit (and of others colors). Rev Bras Frutic. 30(2):566–576.
- Cauz-Santos LA, Munhoz CF, Rodde N, Cauet S, Santos AA, Penha HA, Dornelas MC, Varani AM, Oliveira GCX, Bergès H, et al. 2017. The chloroplast genome of Passiflora edulis (Passifloraceae) assembled from long sequence reads: structural organization and phylogenomic studies in Malpighiales. Front Plant Sci. 8:334.
- Rudnicki M, Oliveira M, V, Pereira T, Reginatto F, Dal-Pizzol F, Fonseca J. 2007. Antioxidant and antiglycation properties of Passiflora alata and Passiflora edulis extracts. Food Chem. 100(2):719–724.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford). 30(9):1312–1313. England).
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones E, Fischer A, Bock R, Greiner S. 2017. GeSeq - Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yang X-L, Hu X-J, Long X-Q. 2019. Characterization of the complete chloroplast genome of a purple passion flower variety ‘Pingtang No.1’ (Passiflora edulia Sims) in China and phylogenetic relationships. Mitochondrial DNA Part B. 4(2):2649–2651.
- Yanqiong C, Lan S-R, Liu Z-J, Zhai J-W. 2019. The complete chloroplast genome sequence of Calanthe delavayi (Orchidaceae), an endemic to China. Mitochondrial DNA Part B. 4(1):1562–1563.
