Abstract
Senna tora and Senna occidentalis, members of Fabaceae, are medicinally important species for laxatives, diuretics, and anticancer those are native to tropical and subtropical areas and Americas, respectively. In this study, the two complete chloroplast (cp) genome sequences in Senna genus were presented. The complete cp genomes of S. tora and S. occidentalis were 162,426 bp and 159,993 bp in size, respectively. Two cp genomes equally harbored 111 genes, including 77 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Phylogenetic analysis based on protein-coding genes demonstrated that S. tora is mostly related with S. occidentalis.
Senna tora and Senna occidentalis are medicinal plants that are widely distributed in tropical and subtropical areas and Americas, respectively. The various tissues such as seed or leaf of S. tora (foremely Cassia tora) and S. occidentalis (foremely Cassia occidentalis) are being used as medicinal stuff in Ayurveda and Chinese traditional medicine for various alignments, such as liver tonic, constipation, ringworm infection, skin diseases, and others (Patil et al. Citation2004; Pawar and D’mello Citation2011). Senna occidentalis are used for antimalarial, anti-inflammatory, and antihemolytic medicines (Neboh and Ufelle Citation2015). Seeds of both S. tora and S. occidentalis were obtained from National Agrobiodiversity Center, National Institute of Agricultural Sciences, Rural Development Administration, Jeonju 54874, Republic of Korea. Fresh leaves used for sequencing were obtained from the National Agrobiodiversity Center (voucher number: IT89788 for S. tora cv. Myeongyun; voucher number: IT175099 for S. occidentalis; geographic coordinates: 35°49′19ʺN, 127°8′56ʺE). Whole genome sequencing was performed using Illumina Hiseq2500 platform (Illumina, USA) and conducted to assemble using CLC assembly cell package (v. beta 4.2.1, CLC Inc., Denmark) as previously mentioned in Kim et al. (Citation2015). The complete cp genome sequences were assembled and joined from three contigs into a single contig by comparison of Cicer canadensis (GenBank accession number KF856619) as a reference. The draft sequence was confirmed and manually corrected by pair-end read mapping. Gene annotation in the cp genome was conducted using DOGMA program (Wyman et al. Citation2004) and NCBI BLASTN searches.
The complete cp genomes of S. tora and S. occidentalis were circular with 162,426 bp and 159,993 bp in length, respectively, and deposited in the Genbank (accession number NC030193 for S. tora; accession number NC038222 for S. occidentalis). The two Senna cp genomes identically harbor 111 annotated genes, including 77 protein-coding genes, 30 tRNA genes, and 4 rRNA genes.
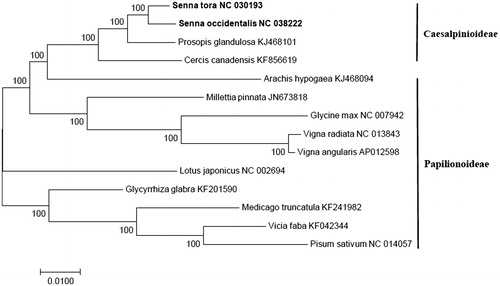
To reveal the phylogenetic relationship of S. tora and S. occidentalis in the Fabaceae, maximum likelihood (ML) phylogeny was analyzed using the protein-coding gene by MEGA 7 program (MEGA Inc., Englewood, NJ) (Tamura et al. Citation2013). As a result, the phylogenetic tree indicated that S. tora was the most related species to S. occidentalis in subfamily Caesalpinioideae (). These complete cp genomes will be helpful for Fabaceae evolution and DNA barcoding studies.
Figure 1. Maximum likelihood (ML) phylogenetic tree constructed based on the protein-coding genes in 14 Fabaceae species, including two Senna species, two subfamily Caesalpinioideae species, and 10 subfamily Papilionoideae species by Mega 7.0 program. Numbers on the node indicate bootstrapping values.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ. 2015. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLOS One. 10(6):e0117159.
- Neboh EE, Ufelle SA. 2015. Myeloprotective activity of crude methanolic leaf extract of Cassia occidentalis in cyclophosphamide-induced bone marrow suppression in Wistar rats. Adv Biomed Res. 4(1):5–8.
- Patil UK, Saraf S, Dixit VK. 2004. Hypolipidemic activity of seeds of Senna tora Linn. J Ethnopharmacol. 90(2–3):249–252.
- Pawar HA, D’mello PM. 2011. Senna tora Linn: an overview. Int J Pharmaceut Sci Res. 2:2286–2291.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
