Abstract
The Basa catfish, Pangasius bocourti, belongs to family Pangasiidae, and is distributed in Mekong and Chao Phraya basins. In this paper, the complete mitochondrial genome of P. bocourti was determined using next-generation sequencing. The whole mitogenome is a typical circular DNA molecule of 16,522 bp and contains 13 protein-coding genes, 22transfer RNA genes, 2 ribosomal RNA genes, and one D-loop region, with the base composition of A 30.9%, G 15.3%, T 26.6%, and C 27.2%. Phylogenetic analysis showed that P. bocourti was the nearest sister to Pangasianodon hypophthalmus. Our whole mitogenome presented here would be useful for further study of P. bocourti.
The Pangasius bocourti belongs to the family Pangasiidae and the species is distributed in Mekong and Chao Phraya basins (Hung et al. Citation1999; Rathod et al. Citation2018;. Limited mitochondrial genome sequences have been reported from family Pangasiidae. Here, we sequenced the complete mitochondrial genome of the adult fish collected from Qingyuan in Guangdong (latitude: 23°34′47.73″N, longitude: 112°53′31.29″), Qingyuan YuShun Fisheries Company, to provide reference information for further study on this species.
The typical specimen were deposited in the Foshan University aquatic museum (P.boco-001). Total genomic DNA was extracted from muscle tissue using the standard phenol-chloroform protocol. Then, the paired-end DNA library with insert size of 400 bp was constructed and sequenced by Illumina X-ten with 150 bp in read length (Zhang et al. Citation2018). The de novo assembly of the mitochondrial genome was assembled by NOVOplasty (Dierckxsens et al. Citation2016).
The complete mitochondrial genome of P. bocourti (GenBank Accession no. MN842723, 16,522 bp) contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 D-loop region. The whole base composition of the mitogenome is showed as follows: A 30.9%, G 15.3%, T 26.6%, and C 27.2%. Total length of the 13 protein-coding genes is 11411 bp, all of which are encoded on the heavy strand except for nad6 in the light strand.
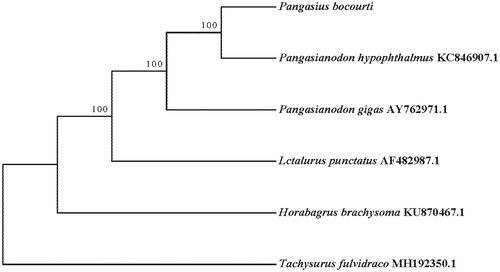
We preformed phylogenetic analysis of Pangasius bocourti species and plus Tachysurus fulvidraco outgroup species, Pangasianodon hypophthalmus, Pangasianodon gigas, Lctalurus punctatus and Horabagrus brachysoma based on 13 protein-coding genes sequences used maximum likelihood method implemented in the RAxML (Silvestro and Michalak Citation2012) (). Pangasianodon hypophthalmus was the nearest sister to Pangasius bocourti. The complete mitochondrial genome of Pangasius bocourti we determined would be useful in systematics and population genetics.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18–e18.
- Hung LT, Tam BM, Cacot P, Lazard J. 1999. Larval rearing of the Mekong catfish, Pangasius bocourti (Pangasiidae, Siluroidei): substitution of Artemia nauplii with live and artificial feed. Aquatic Living Resour. 12(3):229–232.
- Rathod NB, Pagarkar AU, Pujari KH, Shingare PE, Satam SB, Phadke GG, Gaikwad BV. 2018. Status of valuable components from Pangasius: a review. Int J Curr Microbiol App Sci. 7(04):2106–2120.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Zhang Z, Zhang K, Chen S, Zhang Z, Zhang J, You X, Bian C, Xu J, Jia C, Qiang J, et al. 2018. Draft genome of the protandrous Chinese black porgy, Acanthopagrus schlegelii. Gigascience. 7(4):giy012.