Abstract
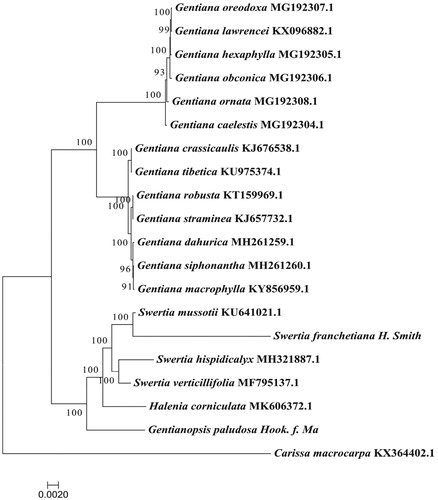
Asone of the original plants of traditional Tibetan folk medicinal ‘Zangyinchen’, Swertia franchetiana (S. franchetiana) was used to treat a variety of ailments. Large amount of fruiting harvesting has led to a sharp decrease and gradual depletion of its resources. As a result, the wild resource is on the verge of extinction, it needs urgent conservation. Here, we report the complete sequence of the chloroplast genome of S. franchetiana. The genome was 153,087 bp in length with 130 genes comprising 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content of G. paludosa chloroplast genome was is 38.17%. Phylogenomic analysis suggested that S. franchetiana formed a clade with Swertia mussotii with high bootstrap values, indicating that the S. franchetiana was closely related to S. mussotii
Swertia franchetiana (S. franchetiana), belonging to the family Gentianaceae, is one of original plants of traditional Tibetan folk medicinal species called ‘Zangyinchen’ (Yang Citation1991). It is mainly distributed in Qinghai, Xizang, Sichuan, and the south of Gansu with high altitude (2300–3800 m) (He et al. Citation1988). It is well known for the treatment of a variety of ailments, including bilious hepatitis, cholecystitis, hepatogenous jaundice, and hypoglycemia (Wang et al. Citation1994, Citation2005; Sun et al. Citation2009). Owing to its therapeutic activities against diseases, the resource demand of S. franchetiana has increased rapidly. In particular, large amount of fruiting harvesting has led to a sharp decrease and gradual depletion of its resources (Liu et al. Citation1996). As a result, its geographical distribution has become increasingly narrow, and the wild resource is on the verge of extinction. So far, the research of S. franchetiana has mainly focused on its chemical composition (Li et al. Citation2008; Liu et al. Citation2008; Sun et al. Citation2009; Li et al. Citation2017), cultivation (Chen et al. Citation2005), and mineral elements (Li et al. Citation2005). Therefore, it is beneficial to understand more genetic information about S. franchetiana to perform conservation, genetic improvement, and sustainable management for this risk species.
We collected a wild individual of S. franchetiana from Xinping village (101°37.556′E, 36°34.460′N, 2510 m), Huangzhong country in Qinghai province of China (Voucher specimen: QHGC20190821, HNWP). We used the following process to sequence the complete chloroplast genome of S. franchetiana: extraction of Genome DNA, fragmentation of DNA, construction of Illumina pair-end library, sequencing, assembling and annotation. The annotated genomic sequence had been submitted to GenBank with the accession number MT188728.
The total length of complete chloroplast genome of S. franchetiana is 153,087 bp, which consisted of a large single-copy region (LSC, 83,220 bp) a small single-copy region (SSC, 18,369 bp) and two inverted repeats (IR, 25,749 bp). The overall CC content is 38.17%. The chloroplast genome includes a total of 130 functional genes including 85 protein-coding genes, 37 tRNA, and 8 rRNA. A total of 18 genes were duplicated in the IR regions including seven tRNA, four rRNA, and seven protein-coding genes. The genome organization, gene/intron content and gene relative positions of the newly sequenced plastid genome were almost identical to other Gentianaceae species.
Phylogenetic tree was reconstructed using the complete chloroplast genomes of S. franchetiana and 18 other species from Gentianaceae, and Carissa macrocarpa (Apocynaceae) was used as an out-group. Maximum-likelihood (ML) analysis showed that S. franchetiana formed a clade with Swertia mussotii with high bootstrap values (), indicating that the S. franchetiana was closely related to S. mussotii.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chen GC, Lu XF, Sun J, Zhou GY, Li JP. 2005. Study on introduction and cultivation of Swertia franchetiana. China J Chin Mater Med. 30(24):1957–1958.
- He TN, Liu SW, Wu QR. 1988. Flora of China. Beijing, China: Science press, p. 402.
- Li HM, Chen T, Sun J, Wang WQ, Li YL. 2017. Separation of six xanthones from Swertia franchetiana by high-speed countercurrent chromatography. J Sep Sci. 40(11):2515–2521.
- Li TC, Chen GC, Sun J, Zhou GY, Li JP. 2005. Study on mineral elements for cultivated Swertia franchetiana H. Smith from Qinghai province. Res Prac Chin Med. 19(4):29–31.
- Li YL, Ding CX, Wang HL, Suo YR, You JM, Chen GC. 2008. Separation and determination of flavone and xanthone glycosides in Tibetan folk medicinal species Swertia mussotii and S. Franchetiana by capillary electrophoresis. J Anal Chem. 63(6):574–579.
- Liu HQ, Liu YR, Zhu ZQ. 1996. Development and protection of medicinal plant resources of Swertia in Qinghai. Chin Tradit Herbal Drugs. 27(2):112.
- Liu YM, Xu Q, Xue XY, Zhang FF, Liang XM. 2008. Two-dimensional LC-MS analysis of components in Swertia franchetiana Smith. J Sep Sci. 31(6–7):935–944.
- Sun YG, Zhang X, Xue XY, Zhang Y, Xiao HB, Liang XM. 2009. Rapid identification of polyphenol C-glycosides from Swertia franchetiana by HPLC-ESI-MS-MS. J Chromatogr Sci. 47(3):190–196.
- Wang JN, Hou CY, Liu YL, Lin LZ, Gil RR, Cordell GA. 1994. Swertifrancheside, an HIV-reverse transcriptase inhibitor and the first flavone-xanthone dimer, from Swertia franchetiana. J Nat Prod. 57(2):211–217.
- Wang LX, Zhang LF, Wang SS, Xiao HB, Liang XM. 2005. Discriminating the xanthones in the extract of Swertia franchetiana by retention parameters. Phytochem Anal. 16:34–38.
- Yang YC. 1991. Book of Tibetan medicine. Xining, China: Qinghai People’s Press.