Abstract
In this paper, we analyzed the complete mitochondrial genome sequence of Ocypode stimpsoni. Whole-genome sequencing was performed using the MGISEQ-2000 platform. The length of the complete mitochondrial genome was 15,557 bp, and data were submitted to NCBI (MN917464). The genome consisted of 67.8% A + T bias, it included 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes, and a putative control region. The phylogenetic tree was constructed based on sequences of 13 PCGs identified by the maximum likelihood (ML) method. Collectively, the results support the original taxonomy assigned, O. stimpsoni belongs to Ocypodidae, Ocypodoidea. In addition, species in Ocypodoidea and Grapsoidea have a reciprocal paraphyly relationship, as demonstrated in previous studies.
Ghost crabs of the genus Ocypode commonly inhabit sandy beaches in tropical and temperate regions (Lucrezi et al. Citation2009). While a total of 25 species have been identified worldwide (Horton et al. Citation2020), the complete mitochondrial genome has only been sequenced for two species, namely O. ceratophthalmus and O. cordimanus (Sung et al. Citation2016; Tan et al. Citation2016). These species are extensively distributed in the Indo-West Pacific region and are more researched than other species (Hughes et al. Citation2014). The distribution of O. stimpsoni in Korea, Japan, and Taiwan has been investigated, and the species has been studied in behavioral and physiological fields; however, genetic research remains limited (Imafuku et al. Citation2001; Tsai and Lin Citation2012). We have, therefore, analyzed the complete mitochondrial genome sequence of O. stimpsoni and compared it with other species with known mitochondrial genome sequence in the infraorder Brachyura. The results may contribute to research on evolution and speciation of Brachyura species.
The specimen was collected on Jeju Island (Korea) in August 2019 (33° 12′ 39.6″ N, 126° 15′ 36.8″ E), preserved in 80% ethanol, and stored in the Ewha Womans University Natural History Museum of Korea with accession number EWNHMAR765. Total DNA was extracted from leg muscle tissue using the MGI Easy DNA Library Prep Kit (MGI, China). Whole-genome sequencing was performed using the MGISEQ-2000 platform. The mitochondrial genome was constructed using MITObim v1.9.1 (Hahn et al. Citation2013) and MITOS (Bernt et al. Citation2013). Annotation was performed using Geneious Prime 2019.2.1 (Kearse et al. Citation2012). Alignment of O. stimpsoni, 59 Brachyura species genome data, and two Anomura species (the outgroup) was performed using ClustalW (Thompson et al. Citation2003). The phylogenetic tree was constructed based on sequences of 13 protein-coding genes (PCGs) identified by the maximum likelihood (ML) method using IQ-TREE (Nguyen et al. Citation2015). The GTR + F + R7 model was identified as the best-fit model for the data, using ModelFinder (Kalyaanamoorthy et al. Citation2017) with 100 bootstrap replicates.
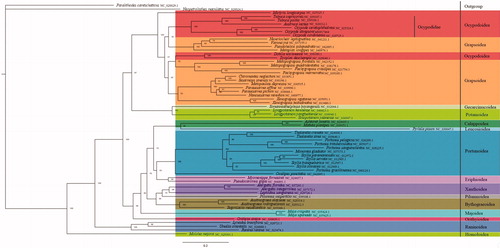
The length of the complete mitochondrial genome was 15,557 bp, and data were submitted to NCBI (MN917464). The genome consisted of 67.8% A + T bias (A = 33.7%, C = 20.8%, G = 11.4% and T = 34.1%). In addition, it included 13 PCGs, two rRNA genes, 22 tRNA genes, and a putative control region consisting of 678 bp. The PCGs used diverse start codons, including ATG (ATP8, COX1, COX2, COX3, CYTB, NAD1, NAD2, NAD4, NAD4L, and NAD5), ATT (ATP6 and NAD6), and ATC (NAD3). We reconstructed the phylogenetic tree using the ML method (). O. stimpsoni clustered with O. ceratophthalmus and O. cordimanus in the genus Ocypode, with high nodal value, and was in the same group as Austruca lactea and Tubuca capriocornis, which belong to the family Ocypodidae. Collectively, the results support the original taxonomy assigned, Ocypode stimpsoni belongs to Ocypodidae, Ocypodoidea. In addition, species in Ocypodoidea and Grapsoidea have a reciprocal paraphyly relationship, as demonstrated in previous studies (Chen et al. Citation2018; Yang et al. Citation2019).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen J, Xing Y, Yao W, Zhang C, Zhang Z, Jiang G, Ding Z. 2018. Characterization of four new mitogenomes from Ocypodoidea & Grapsoidea, and phylomitogenomic insights into thoracotreme evolution. Gene. 675:27–35.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Horton T, et al. 2020. World Register of Marine Species. [accessed 2020 March 02]. Available from: http://www.marinespecies.org. at VLIZ.
- Hughes RN, Hughes DJ, Smith IP. 2014. The ecology of ghost crabs. Oceanogr Mar Biol: An Annual Review. 52:201–256.
- Imafuku M, Habu E, Nakajima H. 2001. Analysis of waving and sound‐production display in the ghost crab, Ocypode stimpsoni. Mar Freshw Behav Phy. 34(4):197–211.
- Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lucrezi S, Schlacher TA, Walker S. 2009. Monitoring human impacts on sandy shore ecosystems: a test of ghost crabs (Ocypode spp.) as biological indicators on an urban beach. Environ Monit Assess. 152(1-4):413–424.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Ortmann AE. 1897. Carcinologische Studien. Zool. Jahrb. Abt. Anat. Ontog. Tiere. 10:258–372.
- Sung JM, Lee J, Kim SG, Karagozlu MZ, Kim CB. 2016. Analysis of complete mitochondrial genome of Ocypode cordimanus (Latreille, 1818) (Decapoda, Ocypodidae). Mitochondrial DNA B. 1(1):363–364.
- Tan MH, Gan HM, Lee YP, Austin CM. 2016. The complete mitogenome of the ghost crab Ocypode ceratophthalmus (Pallas, 1772) (Crustacea: Decapoda: Ocypodidae). Mitochondrial DNA A. 27:2123–2124.
- Thompson JD, Gibson TJ, Higgins DG. 2003. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 1:2–3.
- Tsai JR, Lin HC. 2012. A shift to an ion regulatory role by gills of a semi-terrestrial crab, Ocypode stimpsoni. Zool Stud. 51:606–618.
- Yang TT, Liu Y, Xin ZZ, Liu QN, Zhang DZ, Tang BP. 2019. The complete mitochondrial genome of Uca lactea (Ocypodidae, Brachyura) and phylogenetic relationship in Brachyura. Mitochondrial DNA B. 4(1):1319–1320.