Abstract
The Tephritid fly Tephritis femoralis (Diptera: Tephritidae) is a pre-dispersal seed predator species peculiar to the Qinghai-Tibetan Plateau of China. It is a generalist endoparasite, and it is especially harmful to the Asteraceae species in the alpine meadow. The whole mitochondrial genome of T. femoralis is totally 15,117 bp in length, including 13 protein-coding genes (PCGs), 2 ribosome RNA (rRNA), 22 transfer RNA (tRNA), and 1 control region (CR). The mitochondrial genome contains 41.7% for A, 38.2% for T, 11.6% for C, and 8.5% for G in all base composition. The percentage of A + T content is 79.9%. The CR of T. femoralis was shorter than other species in Tephritidae. Our work provides the first complete mitochondrial genome data in the subfamily Tephritinae, which is important to further studies on evolutionary adaptation and phylogenetic analyses.
Tephritis is the third largest genus in the subfamily Tephritinae, with about 170 described species and more than 20 undescribed species over the world (Korneyev and Korneyev Citation2019). Tephritis larvae are usually an endoparasite: female flies puncture developing flowerheads of plants with their ovipositors and deposit eggs, wherein the larvae grow and develop by consuming developing seeds and do not move until they emerge as adults. Tephritis femoralis is a generalist seed-predator species peculiar to the alpine meadows of the Qinghai-Tibetan Plateau. In northwest Sichuan Province of China, it has been found that T. femoralis may infest 17 Asteraceae species. Thus, Tephritis femoralis may have significant effects on the seed production and population regeneration of these Asteraceae plants.
The T. femoralis specimen was collected form a Saussurea nigrescens flowerhead in Hongyuan County (N 32°48′, E 102°33′; altitude ≈ 3500 m), Sichuan Province, China, the eastern part of the Qinghai-Tibetan Plateau. The specimens are deposited in School of Life Sciences, Nanjing University, China (voucher number: NJUHYTf20180720). Genome DNA extracted from 100% ethanolfixed tissues by the standard phenol/chloroform method. The complete mitochondrial genome was sequenced by Applied BioSystems 3730xl DNA Analyzer (Applied Biosystems, Carlsbad, CA) (Liu et al. Citation2018). The PCGs and rRNA of the mitogenome were annotated by the MITOS web server (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). The tRNA of mitogenome was identified by the program tRNAscan-SE 1.21 (http://lowelab.ucsc.edu/tRNAscan-SE/) (Lowe and Eddy Citation1997).
The mitogenome of T. femoralis is 15,117 bp in length (GenBank accession no. MN857172) and encoded 37 genes totally containing 13 protein-coding genes (PCGs), 2 rRNA, 22 tRNA, and a non-coding region (putative control region, CR). The genome composition of T. femoralis is A = 6304 (41.7%), T = 5775 (38.2%), C = 1754 (11.6%), and G = 1284 (8.5%). There were 14 tRNA genes (tRNALeu(TAA), tRNALys, tRNAAsp, tRNAGly, tRNAAla, tRNAArg, tRNAAsn, tRNASer(GCT), tRNAGlu, tRNAThr, tRNASer(TGA), tRNAIle, tRNAMet, tRNATrp) as well as 9 PCGs genes (COX1, COX2, ATP8, ATP6, COX3, ND3, DN6, CYTB, ND2) encoded on the light strand, with the remaining 14 genes (tRNAPhe, tRNAHis, tRNAPro, tRNALeu(TAG), tRNAVal, tRNAGln, tRNACys, tRNATyr, ND1, ND4, ND5, ND4L, 16S rRNA, 12S rRNA) encoded on the heavy strand. Except for COX1 gene started with TTG, the start code of the remaining 12 PCGs used ATN as their start signal. All of the 13 PCGs ended with TAN in the mitochondrial genome. The control region was 538 bp in length, and the content of A + T in this region was 86.2%.
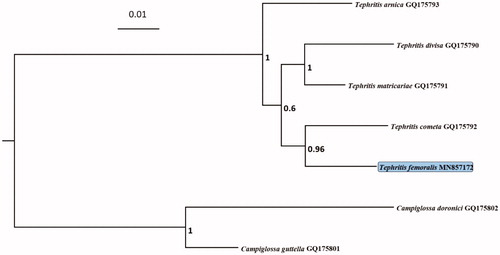
The mitogenome sequence is really limited of the genus Tephritis and even the subfamily Tephritinae. Thus, seven species of the Tribe Tephritini were chosen to reconstruct the phylogenetic tree. The phylogenetic analyses were conducted using the Bayesian inference (BI) in MrBayes v3.1.2 (Ronquist and Huelsenbeck Citation2003). Phylogenetic analysis revealed that T. femoralis had a closer relationship with Tephritis cometa (). The complete mitochondrial genome data of Tephritis femoralis can serve as a basis for further studies on its adaptive evolution and the phylogeny of Tephritid flies.
Figure 1. Phylogenetic relationships of 7 species within the Tribe Tephritini based on the Bayesian inference for combined data set (COI + COII + 16S). The numbers above the branches represent Bayesian posterior probabilities (BI). The number after the species name is the GenBank Accession number. The tree was rooted with the sequences of the outgroup species belonging to the genus Campiglossa.
Acknowledgment
The authors thank Dr Xiaolin Chen from Zoology of Institute, Chinese Academy of Sciences for species identification.
Disclosure statement
No conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Korneyev SV, Korneyev VA. 2019. Revision of the Old World species of the genus Tephritis (Diptera, Tephritidae) with a pair of isolated apical spots. Zootaxa. 4584(1):1–73.
- Liu NY, Wang SY, Yang XJ, Song J, Wu J, Fang J. 2018. The complete mitochondrial genome of Carabus (Damaster) lafossei (Coleoptera: Carabidae). Conserv Genet Resour. 10(2):157–160.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 25(5):955–964.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
