Abstract
The mitochondrial genomes have the potential to be ‘molecular clock’ due to its high mutation rate and low DNA recombination rate. Here, we sequenced the complete mitochondrial genome of three strains of Calonectria ilicicola (anamorph Cylindrocladium parasiticum) isolated from diseased peanut showing symptoms of Cylindrocladium black rot (CBR) and two C. ilicicola strains isolated from diseased soybean showing symptoms of Red crown rot (RCR). The complete mitogenome of C. ilicicola ranged from 41,187 bp to 64,421 bp in length, 22–34 protein-coding genes, 19–20 tRNA genes, 2 rRNA genes, and a control region. Besides, we used a phylogenomic approach to infer evolutionary relationships among 591 mitochondrial genomes that span the diversity of 294 fungal species, including C. ilicicola. Our results underline the potential importance of mitochondrial genomes in comparative genomic analyses and provide a robust evolutionary insight across the tree of life.
The fungus Calonectria ilicicola Boedijin & Reitsma (anamorph: Cylindrocladium parasiticum Crous, Wingfield and Alfennas) is an economically important plant pathogen, which cause Cylindrocladium black rot (CBR) on peanut (Arachis hypogaea L.) and Red crown rot (RCR) on soybean [Glycine max (L.) Merr.] (Gai et al. Citation2017). Calonectria ilicicola is a soil-borne fungal pathogen of global importance, which was first recorded in Georgia, the USA in 1965 and has since spread throughout peanut production areas in the southeastern United States (Bell and Sobers Citation1966; Wright et al. Citation2010). CBR was first recorded on peanut in several counties in Guangdong Province in southern China in 2008, and subsequently observed on peanut in Jiangxi and Fujian Provinces, China, from 2010 to 2012 (Pan et al. Citation2009, Citation2012; Gai et al. Citation2012). RCR was first recorded on soybean in Boluo County, Guangdong Province, China, and subsequently reported on soybean in Tonglu County, Zhejiang Province in eastern China in 2015 (Guan et al. Citation2010; Gao et al. Citation2012; Gai et al. Citation2016).
The mitochondrial genomes have the potential to be ‘molecular clock’ due to its high mutation rate and low DNA recombination rate. In order to understand mitogenomic background and genetic evolution relationship of C. ilicicola, we sequenced the complete mitochondrial genome of three strains isolated from diseased peanut showing symptom of CBR in Boluo County, Huizhou City, Guangdong (23°27′31″N, 114°31′53″E, GDBL02); Longnan County, Ganzhou City, Jiangxi Province (24°38′10″N, 114°45′33″E, JXLN31), and Longyan City, Fujian Province (25°7′23″N, E116°48′7″E, FJLY41), respectively, and two strains isolated from diseased soybean showing symptom of RCR in Boluo County, Huizhou City, Guangdong (23°27′31″No, 114°31′53″E, GDBL60), and Tonglu County, Hangzhou City, Zhejiang Province (29°50′43″N, 119°27′59″E, ZJHZ01), respectively. The taxonomic position of these strains have been identified based on ribosomal DNA internal transcribed spacer (rDNA-ITS), beta-tubulin gene, translation elongation factor 1-a gene sequences, and the pathogenicity has been confirmed by inoculation experiment (Gai et al. Citation2017). The DNA was extracted from the hyphae of C. ilicicola using a fungal DNA extraction kit (OMEGA BioTek, Beijing, China) according to the manufacturer’s instructions. Raw data generated by Illumina HiSeq Xten (Illumina Inc.; San Diego, CA, USA) were subject to de novo assembly by SPAdes version 3.13, followed by 3 rounds of Gap filling with GapCloser version 1.12. The complete mitochondrial genomes were annotated by Mfannot (Beck and Lang Citation2010) and submitted to NCBI GenBank (GDBL02, Genbank accession: MT118656; JXLN31, Genbank accession: MT118659; FJLY41, Genbank accession: MT118655; GDBL60, Genbank accession: MT118657; ZJHZ01, Genbank accession: MT118658). The mitochondrial genome sizes of GDBL02, JXLN31 and FJLY41 are 41,187 bp, 46,105 bp and 39,891 bp, respectively. The mitochondrial genome sizes of GDBL60 and ZJHZ01 are 60,843 bp and 64,421 bp, respectively. The complete mitogenome of C. ilicicola includes 22–34 protein-coding genes, 19–20 tRNA genes, 2 rRNA genes, and a control region. Interestingly, we found that the C. ilicicola strains isolated from diseased soybean showing symptoms of RCR hold larger mitochondrial genome size than C. ilicicola strains isolated from diseased peanut showing symptoms of CBR.
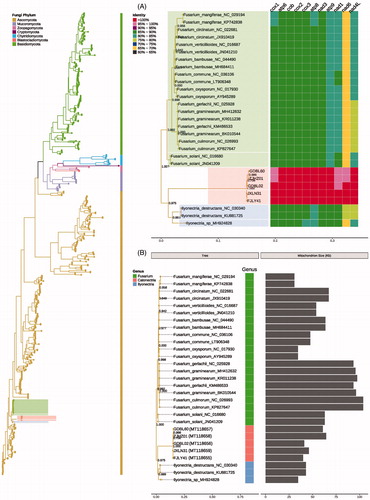
In addition, we used the BLAST-based orthology detector OrthoFinder v2.2.7 with default parameter values to identify orthogroups among all the protein sequences of the 591 mitochondrial genomes (Emms and Kelly Citation2019). We constructed sequence alignment of 11 conserved PCGs extracted from 591 mitochondrial genomes that span the diversity of 294 fungal species by MAFFT v7.313 to reveal the phylogenetic relationship of C. ilicicola and relative fungi species (Katoh and Standley Citation2013). We reconstructed the complete mitochondrial genome-based phylogenomic tree of five C. ilicicola strains and 586 fungal mitochondrial genomes using FastTree v2.1.10, and the tree was tested with 1000 Bootstrap replications (Price et al. Citation2010). The phylogenomic tree is shown in and we can see from the figure that all 591 fungal strains were divided into different clades in accordance with Phylum classification. We found that the five C. ilicicola strains are closest to the genus Ilyonectria and Fusarium, which were located in a clade in the Phylum of Ascomycota (). This study can provide the basis for the genetic evolution of C. ilicicola and underline the potential importance of mitochondrial genomes in comparative genomic analyses, which provide a robust evolutionary insight across the tree of life in the near future.
Acknowledgements
Authors are grateful to Dr. Xinglong Chen and Dr. Dagao Xu for their valuable advice.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Beck N, Lang BF. 2010. Mfannot, organelle genome annotation webserver. [accessed 2020 Mar 28]. http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl
- Bell DK, Sobers EK. 1966. A peg, pod, and root necrosis of pea-nuts caused by a species of Calonectria. Phytopathology. 56:1361–1364.
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):1–14.
- Gai Y, Deng Q, Chen X, Guan M, Xiao X, Xu D, Deng M, Pan R. 2017. Phylogenetic diversity of Calonectria ilicicola causing Cylindrocladium black rot of peanut and red crown rot of soybean in southern China. J Gen Plant Pathol. 83(5):273–282.
- Gai Y, Deng Q, Pan R, Chen X, Deng M. 2012. First report of Cylindrocladium black rot of peanut caused by Cylindrocladium parasiticum (teleomorph Calonectria ilicicola) in Jiangxi Province, China. Plant Dis. 96(4):586–586.
- Gai Y, Huang Z, Chen H, et al. 2016. Identification of red crown rot of soybean caused by Calonectria ilicicola in Zhejiang Province, China. Soybean Sci. 35(6):986–991.
- Gao X, Lu X, Wu M, Zhang H, Pan R, Tian J, Li S, Liao H. 2012. Co-inoculation with rhizobia and AMF inhibited soybean red crown rot: from field study to plant defense-related gene expression analysis. PLOS One. 7(3):e33977.
- Guan M, Pan R, Gao X, Xu D, Deng Q, Deng M. 2010. First report of red crown rot caused by Cylindrocladium parasiticum on soybean in Guangdong, Southern China. Plant Dis. 94(4):485–485.
- Katoh K, Standley DM. 2013. Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Pan R, Deng Q, Xu D, Ji C, Deng M, Chen W. 2012. First report of peanut Cylindrocladium black rot caused by Cylindrocladium parasiticum in Fujian Province, eastern China. Plant Dis. 96(4):583–583.
- Pan R, Guan M, Xu D, Gao X, Yan X, Liao H. 2009. Cylindrocladium blackrot caused by Cylindrocladium parasiticum newly reported on peanut in China. Plant Pathol. 58(6):1176–1176.
- Price MN, Dehal PS, Arkin AP. 2010. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLOS One. 5(3):e9490.
- Wright LP, Davis AJ, Wingfield BD, Crous PW, Brenneman T, Wingfield MJ. 2010. Population structure of Cylindrocladium parasiticum infecting peanuts (Arachis hypogaea) in Georgia, USA. Eur J Plant Pathol. 127(2):199–206.