Abstract
Bauhinia variegata L. var. variegata is a popular ornamental tree in the tropical and subtropical region. We herein report and characterize the complete plastid genome of B. variegata var. variegata in an effort to provide genomic resources for genetic utilization. The complete plastome is 155,415 bp in length and contains the typical quadripartite structure, including two inverted repeat (IR) regions of 25,549 bp, a large single-copy (LSC) region of 86,110 bp and a small single-copy (SSC) region of 18,207 bp. 130 genes are annotated, including 85 protein-coding genes, 37 transfer RNA genes, and four unique ribosomal RNA genes. The phylogenetic analysis based on the plastomes from B. variegata var. variegata and 12 previously reported species of Cercidoideae suggested a sister-relationship between B. variegata var. variegata and B. × blakeana with a strong bootstrap value.
Bauhinia variegata L. var. variegata, with large, fragrant flowers and long flowering time, is a well-known ornamental tree of the legume family in tropical and subtropical region. Evidence of morphology and Sanger sequencing suggests that it could be one of the parental species of the Hong Kong Orchid Tree, Bauhinia × blakeana Dunn (Lau et al. Citation2005; Mak et al. Citation2008; Kumari and Meenakshi Citation2013). B. variegata var. variegata and B. × blakeana are comparable in morphology by having five fertile stamens and leaves bifid to 1/3–1/2. Nevertheless B. variegata var. variegata has pink petals and linear legume, which is very different from the completely sterile B. × blakeana with purplish flowers. To better understand the plastid genome characterization of this plant, we generated the complete plastid genome of the species using the method of genome skimming.
The fresh leaf tissues were collected from South China Agricultural University, Guangzhou, China (113.35°E, 23.16°N). Voucher specimens (HN1904242) were deposited in the herbarium of South China Botanical Garden (IBSC). We isolated the whole genomic DNA by a modified CTAB method (Doyle and Doyle Citation1987). We constructed a library for sequencing by fragmenting the total genomic DNA into 300–500 bp in length following the manufacturer’s manual (Illumina). Paired-end (PE) sequencing was conducted on the Illumina HiSeq X-Ten instrument at Beijing Genomics Institute (BGI) in Wuhan, China. We used GetOrganelle pipeline (Bankevich et al. Citation2012; Langmead and Salzberg Citation2012; Wick et al. Citation2015; Jin et al. Citation2018) to assemble the plastome from clean sequencing reads. We employed Geneious (Kearse et al. Citation2012) and Plastid Genome Annotator (PGA) (Qu et al. Citation2019) to verify the accuracy of the assembly and to annotate the plastome. The annotated plastome has been deposited in GenBank (accession number: MT176420).
To reconstruct the phylogenetic position of B. variegata var. variegata, we included 13 plastid genomes in previous publications and unpublished data in GenBank () (Sabir et al. Citation2014; Wang et al., Citation2017, Citation2018; Gu et al. Citation2019). We aligned the data matrix using MAFFT (Katoh and Standley Citation2013) as implemented in Geneious with default parameters. The maximum likelihood tree was built in IQ-tree (Trifinopoulos et al. Citation2016) using models recommended by ModelFinder (Kalyaanamoorthy et al. Citation2017) based on a data matrix of a concatenation of 77 CDS genes. The branch supports were estimated using 1000 replicates of bootstrap.
Figure 1. The maximum-likelihood (ML) phylogenetic tree based on a concatenation of 77 protein-coding genes. Numbers on the branches are bootstrap support values.
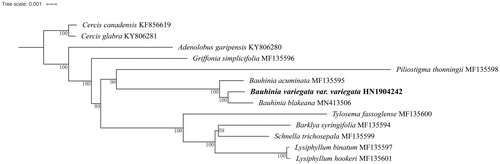
The complete plastid genome of B. variegata var. variegata was 155,415 bp in length and showed a typical quadripartite structure: a large single copy (LSC) region of 86,110 bp and a small single copy (SSC) region of 18,207 bp, respectively. These two regions were separated by two inverted repeat regions (IRa and IRb), each of 25,549 bp in length. A total of 130 functional genes were recovered, consisting of 85 protein-coding genes, 37 tRNA genes and four unique rRNA genes. The overall GC content was 36.5%. The phylogenetic analysis suggested that B. variegata var. variegata and B. × blakeana form a strongly supported clade, which is sister to B. acuminata L. with a strong bootstrap support value (). The plastid genome of B. variegata var. variegata reported here could be helpful for answering questions regarding genetic diversity in Cercidoideae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Lau CPY, Ramsden L, Saunders RMK. 2005. Hybrid origin of Bauhinia blakeana (Leguminosae- Caesalpinioideae), inferred using morphological, reproductive, and molecular data. Am J Bot. 92(3):525–533.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Gu SR, Lai Q, Zeng QB, Tu TY, Zhang DX. 2019. The complete plastid genome of Hong Kong Orchid Tree, Bauhinia × blakeana Dunn (Leguminosae). Mitochondr DNA B. 4(2):3454–3455.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv.256479. DOI:10.1101/256479
- Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumari NS, Meenakshi S. 2013. Investigation of Bauhinia blakeana Dunn. A hybrid species. Biosci Disc. 4(1):20–24.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie2. Nat Methods. 9(4):357– 357.
- Mak CY, Cheung KS, Yip PY, Kwan HS. 2008. Molecular evidence for the hybrid origin of Bauhinia blakeana (Caesalpinioideae). J Integr Plant Biol. 50(1):111–118.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Sabir J, Schwarz EN, Ellison N, Zhang J, Baeshen NA, Mutwakil M, Jansen RK, Ruhlman TA. 2014. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol J. 12(6):743–754.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Bui QM. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Wang YH, Wang H, Yi TS, Wang YH. 2017. The complete chloroplast genomes of Adenolobus garipensis and Cercis glabra (Cercidoideae, Fabaceae). Conserv Genet Resour. 9(4):635–638.
- Wang YH, Wicke S, Wang H, Jin JJ, Chen SY, Zhang SD, Li DZ, Yi TS. 2018. Plastid genome evolution in the early-diverging Legume subfamily Cercidoideae (Fabaceae). Front Plant Sci. 9:138.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
