Abstract
Wolffia brasiliensis, a species of duckweed, is an aquatic plant belonging to the family Araceae. In this study, the complete chloroplast genome of W. brasiliensis was assembled from the whole genome Illumina sequencing data. The chloroplast genome is 168,514 bp in length, which contained one large single copy (LSC; 90,790 bp) and one small single copy (SSC; 13,930 bp) separated by two inverted repeat (IR) regions of 31,897 bp. It encodes 4 rRNA, 29 tRNA, and 78 protein-coding genes, with 20 double copies. Total GC content is 36.24%. Bayesian phylogenetic analysis indicated that W. brasiliensis occupied the ambiguous phylogenetic position between Wolffiella and Wolffia.
Wolffia brasiliensis (duckweed), a species of the Lemnoideae (duckweed subfamily), is an aquatic floating monocot plant (Hoang et al. Citation2019). It is distributed in North and South America, where it grows mostly on surfaces of calm water, such as ponds. It is a very small plant (about 1 mm in size), with no stems and roots, only spherical fronds (Les et al. Citation1997). Species of the Lemnoideae, including the Wolffia, lack the morphological characters for taxonomic comparison and they require molecular data for phylogenetic relationship analysis (Tippery et al. Citation2015). In particular, the comparison of the complete chloroplast genome provides useful information for the reconstruction of taxonomic locations and for verification of phylogenetic relationships (Mardanov et al. Citation2008). Although the taxonomy of Lemnoideae has been studied for a long time (Les et al. Citation2002), the position of W. brasiliensis remains an issue (Les et al. Citation2002; Wang et al. Citation2010; Tippery et al. Citation2015). The complete chloroplast genome will give insights to solve the taxonomic problems of W. brasiliensis.
For its phylogenetic research, in this study, the complete chloroplast genome of the W. brasiliensis was assembled from whole genome data. Plant material was obtained from the Rutgers Duckweed Stock Cooperative at Rutgers University, New Brunswick, NJ, USA (RDSC; http://www.ruduckweed.org/). Total genomic DNA of W. brasiliensis (strain 7710 from Dajabon, Restauracion in Dominican Republic; 19°18′48″N, 71°41′28″W) was manually extracted using cetyltrimethylammonium-bromide (CTAB). The genomic sequencing was performed by Illumina Hiseq, and the complete chloroplast genome was de novo assembled in Novoplasty v.2.7.2 (Dierckxsens et al. Citation2017). Ribulose 1,5-bisphosphate (RuBP) of W. australiana was used as a seed for the assembly. The assembled chloroplast genome was annotated using the GeSeq (Tillich et al. Citation2017), and the complete chloroplast genome was submitted to GenBank with the accession number MN850406.
The chloroplast genome size of W. brasiliensis is 168,514 bp, with one large single-copy (LSC, 90,790 bp), one small single-copy (SSC, 13,930 bp) and two inverted repeat (IR) regions (31,897 bp each). A total of 111 genes are composed of 78 protein coding, 29 tRNA, and 4 rRNA, of which 20 are double copies. The total GC content is 36.24% and each value of LSC, SSC, and IR is 34.33%, 31.52%, and 39.98%, respectively.
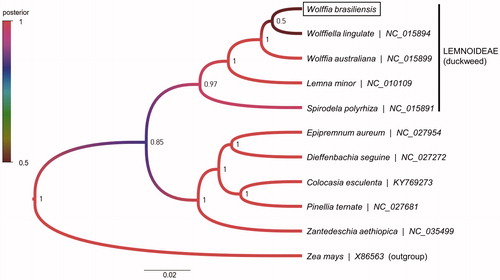
To determine the phylogenetic position of W. brasiliensis, a Bayesian phylogenetic analysis was performed using the BEAST v1.10.4 (Drummond and Rambaut Citation2007). The phylogenetic tree was produced using species in the family Araceae by FigTree v1.4.4 (). As a result, W. brasiliensis was in the Lemnoideae but classified as Wolffiella, not Wolffia. The node posterior between W. brasiliensis and Wolffiella lingulate was as low as 0.5. Suggesting the classification of W. brasiliensis is still ambiguous even with whole chloroplast genome information. The unexpected classification of W. brasiliensis was consistent with other studies (Les et al. Citation2002; Wang et al. Citation2010; Tippery et al. Citation2015). This unclear classification may need to be validated by additional data (such as nuclear DNA) and additional W. brasiliensis accessions.
Figure 1. Bayesian phylogenetic tree of family Araceae from chloroplast DNA data (56 protein-coding genes) by BEAST v1.10.4. Nodal support values are given as Bayesian posterior probability. The CpREV + G + I model was the best-fit model of protein evolution as suggested by ProtTest v3.4.2 (Darriba et al. Citation2011). The position of Wolffia brasiliensis (Genbank accession no. MN850406) is shown in a box.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 27(8):1164–1165.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucl Acids Res. 45:e18.
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 7(1):214.
- Hoang PTN, Schubert V, Meister A, Fuchs J, Schubert I. 2019. Variation in genome size, cell and nucleus volume, chromosome number and rDNA loci among duckweeds. Sci Rep. 9(1):1–13.
- Les DH, Crawford DJ, Landolt E, Gabel JD, Kimball RT. 2002. Phylogeny and systematics of Lemnaceae, the duckweed family. Syst Bot. 27:221–240.
- Les DH, Landolt E, Crawford DJ. 1997. Systematics of the Lemnaceae (duckweeds): inferences from micromolecular and morphological data. Plant Syst Evol. 204(3–4):161–177.
- Mardanov AV, Ravin NV, Kuznetsov BB, Samigullin TH, Antonov AS, Kolganova TV, Skyabin KG. 2008. Complete sequence of the duckweed (Lemna minor) chloroplast genome: structural organization and phylogenetic relationships to other angiosperms. J Mol Evol. 66(6):555–564.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucl Acids Res. 45(W1):W6–W11.
- Tippery NP, Les DH, Crawford DJ. 2015. Evaluation of phylogenetic relationships in Lemnaceae using nuclear ribosomal data. Plant Biol J. 17(Suppl 1):50–58.
- Wang W, Wu Y, Yan Y, Ermakova M, Kerstetter R, Messing J. 2010. DNA barcoding of the Lemnaceae, a family of aquatic monocots. BMC Plant Biol. 10(1):205.
