Abstract
Cyclocodon lancifolius (Roxb.) Kurz is a popular food and traditional folk medicine with highly economic value in China. The complete chloroplast (cp) genome of C. lancifolius was 167,026 bp in length, which contained a large single-copy (LSC) region of 84,648 bp, a small single-copy (SSC) region of 8044 bp, and a pair of inverted repeats (IRs) region of 37,167 bp. The overall GC content was 38.12%. The genome harbored 24 genes, including 12 protein-coding genes, 8 tRNA genes, and 4 ribosomal RNA genes. Phylogenetic analysis indicated that C. lancifolius is closely related to the species of Codonopsis minima and Platycodon grandiflorus.
Cyclocodon lancifolius (Roxb.) Kurz is a perennial herb that belongs to the family of Campanulaceae, and commonly known as Hong Guo Ginseng. It is widely distributed in China, India, Philippines, Vietnam, Cambodia, Myanmar, and Sikkim (Hong et al. Citation2011), C. lancifolius is not only as food but also as traditional Yi folk medicine. The root of C. lancifolius is used to treat trauma, qi deficiency, intestinal colic, tuberculosis cough, scrofula, hernia, etc. (Nanjing Citation2014). Its fruits is rich in polysaccharides, proanthocyanidins, and flavonoids; extracts from the fruits have several pharmacological actions such as radical scavenging, anti-aging, etc. (Wang et al. Citation2014; Li et al. Citation2015). Therefore, C. lancifolius has huge potential medicinal and economic value. However, there have been no complete chloroplast (cp) genomic studies on C. lancifolius. Herein, we assembled and characterized the cp genome of C. lancifolius for the first time, and its phylogenetic analysis is also investigated which will provide informatics data for the phylogeny of genus Cyclocodon.
The fresh leaves of C. lancifolius were collected from the Cangshan mountain (25°68′04″N, 100°15′96″E) of Dali counties, Yunnan province, China. The voucher specimen was deposited in the herbarium of Dali University (LJ2020010501). Total DNA was extracted using the DNeasy plant mini kit (QIAGEN, Hilden, Germany), and sequenced by Illumina NovaSeq system (San Diego, CA). Approximately, 7.20 Gb of raw data (48,026,480 reads) were assembled by NOVOPlasty (Park et al. Citation2019; Liu et al. Citation2020), and the assembled cp genome was annotated by GeSeq with default sets (Michael et al. Citation2017; Qian et al. Citation2020). The annotated cp genome was submitted to the GenBank with the accession number of MT218569.
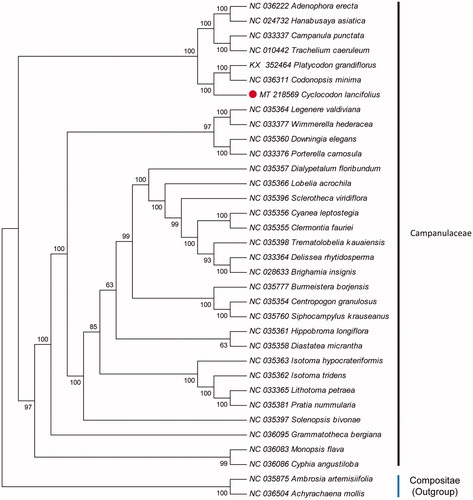
The cp genome of C. lancifolius was 167,026 bp in length with a typical quadripartite structure of angiosperms, containing a large single copy (LSC) region of 84,648 bp, a small single copy (SSC) region of 8044 bp, and a pair of inverted repeats (IRs) region of 37,167 bp. The genome comprises of 24 genes, including 12 protein coding genes, eight tRNA genes, and four ribosomal RNA genes. The overall GC content was 38.12%. To investigate the phylogenetic relationship of C. lancifolius, a total of 31 cp genome sequences from related species in Campanulaceae were downloaded from the NCBI database, and were aligned using MAFFT v7.307 (Katoh and Standley Citation2013). A neighbor-joining (NJ) tree with 1000 bootstrap replicates was inferred using MEGA version 6.0 (Tamura et al. Citation2013), with Ambrosia artemisiifolia (NC 035875) and Achyrachaena mollis (NC 036504) as outgroup. The phylogenetic analysis showed that C. lancifolius was closely related to the species of Codonopsis minima and Platycodon grandiflorus (). The cp genome of C. lancifolius will provide a useful resource for the conservation genetics of this species as well as for phylogenetic studies of Cyclocodon genus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Hong DY, Ge S, Lammers TG, Klein LL. 2011. Flora of China. Vol. 19. Beijing, China: Science Press.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li B, Li G, Chen GX, Li H, Liang JP, Chen Y. 2015. Extraction and composition of polysaccharide from the fruits of Campnumoea lancifolia. Subtrop Plant Sci. 1(44):13–17.
- Liu TX, Qian J, Du ZF, Wang J, Duan BZ. 2020. Complete genome and phylogenetic analysis of Agrimonia pilosa Ldb. Mitochondrial DNA B. 5(2):1435–1436.
- Michael T, Pascal L, Tommaso P, Ue S, Axel F, Ralph B, G S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Nanjing U. 2014. Dictionary of traditional Chinese Medicine. Shanghai: Shanghai Scientific and Technical Publishers.
- Park J, Choi YG, Yun N, Xi H, Min J, Kim Y, Oh S-H. 2019. The complete chloroplast genome sequence of Viburnum erosum (Adoxaceae). Mitochondrial DNA B. 4(2):3278–3279.
- Qian J, Du Z, Jiang Y, Duan B. 2020. The complete chloroplast genome sequence of Althaea rosea (L.) Cavan. (Malvaceae) and its phylogenetic analysis. Mitochondrial DNA B. 5(2):1433–1434.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Wang XJ, Chen LH, Mo YT. 2014. Cooperative antioxidant effects of flavonoids from Hong Guo ginseng and Vc. Food Ferment Ind. 12(40):111–115.