Abstract
We sequenced and annotated the complete mitochondrial genome (mitogenome) of Bactrocera scutellata (Diptera: Tephritidae), which is an economically important pest in many area of East Asia. This mitogenome is 15,850 bp in length with an A + T content of 72.99%, and contains 37 typical animal mitochondrial genes that are arranged in the same order as that of the inferred ancestral insects. All protein-coding genes (PCGs) start with a typical ATN codon, with the exception of cox1, which begins with TCG. Ten PCGs stop with termination codon TAA or TAG, whereas cox1, nad1 and nad5 have single T as the incomplete stop codon. All of the transfer RNA genes present the typical clover leaf secondary structure excepting trnS1 (AGN) without a DHU-arm. The A + T-rich region (945 bp in length with 84.66% A + T content) is located between rrnS and trnI, and contains a 20 bp poly-T stretch and 21 bp poly-A stretch. Excepting the control region, the longest intergenic spacer locates between trnR and trnN which is 56 bp long with an excessive high A + T content (89.29%). Compared with other two mitogenomes of B. scutellata revealed that the mutation rates of mitochondrial genes were quite different. For population genetic studies of this species, we recommend using the cox1 or nad5 genes.
Fruit flies (Diptera: Tephritidae) are among the most economically important pest species in the world, attacking a wide range of fruits and fleshy vegetables throughout tropical and sub-tropical areas (Vargas et al. Citation2015). It is estimated that Tephritidae contains more than 4257 species in 471 genera worldwide (Thompson Citation1998), but to date, very limited mitogenomes have been reported. The striped fruit fly, Bactrocera (Zeugodacus) scutellata (Hendel) (Diptera: Tephritidae) is a vital quarantine pest that causes damage to Cucurbitaceae plants and has been found in China, Korea, Bhutan, Vietnam, Thailand and Japan (Ryukus) (Kim et al. Citation2010; Vargas et al. Citation2015).
The mitogenome of B. scutellata (collected from Beibei District, Chongqing, China and the voucher specimen was deposited at the Insect Herbarium of College of Plant Protection, Southwest University under the accession number Di-14-356) was amplified with 2 pairs of primers (available from corresponding author on request) and sequenced in both directions. It (GenBank accession number, KT159731) was 15850 bp in length and with the same number, order and orientation of genes as that of the inferred ancestral insects (Boore Citation1999). Except the A + T-rich control region, 157 nucleotides ranged from 1 to 56 bp distributed in 15 intergenic spacers. Gene overlaps were 32 bp ranged from 1 to 8 bp distributed in 9 locations with three relatively long overlaps (trnW-trnC, atp8–atp6 and nad4L–nad4) which highly conserved in all sequenced species of Tephritidae except two nucleotides diffterence occurred in overlap of trnW-trnC of Procecidochares utilis.
Phylogenetic analysis indicated that B. scutellata belonged to the subgenus Zeugodacus of Bactrocera, and was closest to B. diaphora (). Since other two mitogenomes of B. scutellata had been sequenced (Liu et al. Citation2017), we analyzed their differences. The results revealed that 121 differential sites (4 in rrnL, 1 in rrnS, 3 in tRNA genes, 8 in AT-rich region, 2 in intergenic regions and 103 in PCGs) were found and the overall substitution rate was 0.77%. The ratio of nucleotide conversion to transversion reached more than 8 times (108/13). For transversions, except only one occurred between C and G, all other occurred between A and T (8 sites) and between A and C (4 sites). Although 103 substitutions located in PCGs, only four substitutions caused amino acid change (S74G in cox1, N10D in atp6, I91M in nad3 and S514G in nad5). Interestingly, none of the substitution site occurred in the second codon position (91 and 12 occurred in first and third codon position respectively).
Figure 1. Phylogenetic tree was constructed using Bayesian inference and Neighbor-joining method based on the complete mitogenome sequences of the species of Tephritidae. Values at each node indicated percentage Bayesian posterior probabilities and the bootstrap percentages from 1000 replicates. The GenBank accession numbers of species showed in the parentheses. The subgenera of Bactrocera represented on the right side of the tree.
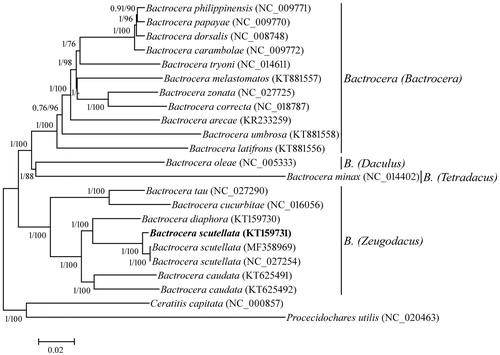
Overall, mutation rates between mitochondrial genes of B. scutellata were significantly different. The mutation rate of PCGs (0.92%) was higher than that of rRNA genes (0.24%) and tRNA genes (0.20%). Among the 13 PCGs, the five highest mutation rates genes were nad3 (1.412%), atp6 (1.18%), cox1 (1.17%), nad5 (1.11%) and cox3 (1.01%), while other PCGs were less than 1% (The atp8 gene even had no mutation). Considering the length of these genes, we suggested that cox1 and nad5 were more suitable for population genetic studies of this species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Kim YP, Jeon SW, Lee SG, Kim KH, Choi NJ, Hwang CY. 2010. Seasonal occurrence and damage of Bactrocera scutellata (Diptera: Tephritidae) in Jeonbuk Province. Kor J Appl Entomol. 49(4):299–304.
- Liu JH, Jia PF, Liu LL, Wang QM, Zhang YX, Ruan SQ. 2017. Complete mitochondrial genome of stripped fruit fly, Bactrocera (Zeugodacus) scutellata (Diptera: Tephritidae) from Anshun, Southwest China. Mitochondrial DNA Part B. 2(2):387–2388.
- Thompson FC. 1998. Fruit fly expert identification system and systematic information Database. Leiden: North American Dipterists’ Society. Backhuys Publishers. p. 524.
- Vargas RI, Piñero JC, Leblanc L. 2015. An overview of pest species of Bactrocera fruit flies (Diptera: Tephritidae) and the integration of biopesticides with other biological approaches for their management with a focus on the Pacific region. Insects. 6(2):297–318.
