Abstract
The genus Lonicera Linn. is among the most speciose in the Northern Hemisphere and has been important horticultural plants in China. Due to the lack of efficient molecular markers, although Lonicera has received the most extensive taxonomic evaluation historically, species discrimination within the genus is still quite difficult to this day. In this study, we characterized the complete chloroplast (cp) genome of L. korolkowii. It was 154,664 bp in length, comprising a pair of inverted repeat (IR) regions (23,735 bp) separated by a large single-copy (LSC) region (88,387 bp) and a small single-copy (SSC) region (18,745 bp). Phylogenetic analysis supported the monophyly of Caprifoliaceae, Adoxaceae and Lonicera, and suggested that Lonicera was closely related to Triosteum pinnatifidum and Heptacodium miconioides, L. korolkowii was related to L. sachalinensis. The complete cp genome of L. korolkowii will contribute to further studies on population genetics, phylogeny and conservation biology in Lonicera.
The genus Lonicera is among the most speciose in the Northern Hemisphere (Hsu and Wang Citation1988). Although Lonicera has received the most extensive taxonomic evaluation historically, species discrimination within the genus is still quite difficult to this day (Theis et al. Citation2008; He et al. Citation2017). Recently, molecular barcodes based on the complete cp genomes have shown great potential for species identification, especially between closely related taxa (Yao et al. Citation2010). In this study, we present the complete genome of L. korolkowii for the first time, which will be beneficial for DNA barcoding studies in Lonicera.
Fresh leaf material was collected from Nanjing Botanical Garden (N 32°03′16.85″, E 118°49′52.06″) and the voucher specimen was deposited in Henan Agricultural University Herbarium (CYN20190522). Total genomic DNA was extracted and sequenced on the Illumina Hiseq2500 Platform at Jinweizhi Biotechnology Institute (Suzhou, China). Firstly, we filtered and assembled the complete genome using CLC Genomics Workbench (http://www.clcbio.com). After that, the draft genome was annotated using GENEIOUS v11.1.4 (http://www.geneious.com). Start/stop codons and the exon/intron boundaries of genes were confirmed according to the cp genome of L. japonica (NC_026839.1). In addition, we used tRNAscan-SE v1.21 (Schattner et al. Citation2005) to further verify the tRNA boundaries with default settings. Then, the online program OrganellarGenome DRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html; Lohse et al. Citation2013) was used to draw the physical map of the cp genome. Finally, to get more knowledge about the phylogenetic position of L. korolkowii, we chose 8 plant species representing 8 genera of Caprifoliaceae, and 14 taxa from Lonicera to reconstruct phylogenetic tree using one species each for Adoxa moschatellina, Tetradoxa omeiensis, Sinadoxa corydalifolia, Viburnum utile, Viburnum betulifolium and Sambucus williamsii as outgroups with PhyML v3.0 (Guindon et al. Citation2010).
The full length of the cp genome of L. korolkowii was 154,664 bp with 38.4% GC content (GenBank accession number: MN823073). Its quadripartite structure was composed of a LSC region (88,387 bp), a SSC region (18,745 bp), and a pair of inverted repeated regions (IRs; 23,735 bp). The genome encoded a set of 122 genes, including 81 protein-coding genes, 31 transfer RNA (tRNA) genes, 8 ribosomal RNA (rRNA) genes and two pseudogenes (accD and petB). Of those genes, 9 contained one intron and 4 contained 2 introns, and it is noteworthy that 14 genes were completely duplicated in the IR regions. The genome organization, gene content, and the relative positions of the 122 genes of L. korolkowii were almost identical to those of L. japonica (He et al. Citation2017)
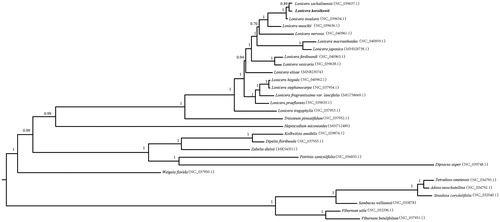
The phylogenetic tree showed all the species of Caprifoliaceae and Adoxaceae formed two distinct monophyletic clades with both 100% bootstrap values (). Within Caprifoliaceae, L. korolkowii together with the other 14 plant species in Lonicera were also clustered into a monophyletic clade (100% bootstrap value), and additionally, our results indicated that Lonicera was closely related to Triosteum pinnatifidum and Heptacodium miconioides, L. korolkowii was related to L. sachalinensis. These results are similar to previous analyses based on various molecular data sets (Fan et al. Citation2018; Wang et al. Citation2020).
Acknowledgments
The authors thank Mr. Tian-lei Xie from Central South University of Forestry and Technology, Dr. Yan-yan Liu, Dr. Jia-mei Li and Cui-jie Sun from Henan Agricultural University for collecting plant materials in China.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Fan WB, Wu Y, Yang J, Shahzad K, Li ZH. 2018. Comparative chloroplast genomics of Dipsacales species: insights into sequence variation, adaptive evolution, and phylogenetic relationships. Front Plant Sci. 9:689.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyMl 3.0. Systematic Biol. 59(3):307–321.
- He L, Qian J, Li XW, Sun ZY, Xu XL, Chen SL. 2017. Complete chloroplast genome of medicinal plant Lonicera japonica: genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules. 22(2):249.
- Hsu P, Wang H. 1988. Lonicera. In: P. Hsu., H. Wang and J. Hu, editors. Flora Republicae Popularis Sinicae. Beijing: Science Press; p. 153–257.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellargenome DRAW: a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:575–581.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:686–689.
- Theis N, Donoghue MJ, Li JH. 2008. Phylogenetics of the Caprifolieae and Lonicera (Dipsacales) based on nuclear and chloroplast DNA sequences. Issn: 0363-6445. 33(4):776–783.
- Wang H-X, Liu H, Moore MJ, Landrein S, Liu B, Zhu Z-X, Wang H-F. 2020. Plastid phylogenomic insights into the evolution of the Caprifoliaceae s.l. (Dipsacales). Mol Phylogenet E. 142:106641.
- Yao H, Song J, Liu C, Luo K, Han J, Li Y, Pang X, Xu H, Zhu Y, Xiao P, et al. 2010. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One. 5(10):e13102.