Abstract
Senecio scandens Buch.-Ham. is a crucial source of Chinese traditional medicine with antibacterial properties. In this study, we report the complete chloroplast genome sequence of S. scandens. The assembled chloroplast genome was 150,729 bp in length, containing two inverted repeated (IR) regions of 24,455 bp each, a large single-copy (LSC) region of 83,984 bp and a small single-copy (SSC) region of 17,835 bp. The genome encodes 133 genes consisting of 89 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The overall GC content of S. scandens is 37.4%, with the highest GC content of 43% in the IR region. A total of 38 simple sequence repeats are identified in the cp genome of S. scandens. Phylogenetic analysis demonstrated a sister relationship between S. scandens and Pericallis hybrida, indicating further revisions for the genus Senecio. This work provides basic genetic resources for investigating the evolutionary status and population genetics diversities for this medicinal species.
Senecio scandens Buch.-Ham. is a medicinal plant from family Asteraceae, which has been recorded as Senecionis Scandentis Herba (Qianliguang) in Chinese Pharmacopeia. It has a climbing woody stem and usually grows on hills, mountains, woods and roadsides. Senecio scandens is widely used as an ingredient to produce hundreds of medicinal plantal preparations with various activities (Wang et al. Citation2013). Because of similar morphological characteristics, several species are easily confused with S. scandens. However, significant differences were found on the chemical markers even between the two species from the genus Senecio (Xiong et al. Citation2014). It is necessary to develop an effective molecular identification strategy for S. scandens to ensure the safety of clinical application. The aim of this study was to analyze the chloroplast genome sequence of S. scandens, which could provide essential information for molecular marker development and analysis of phylogenetic status of this species.
The sample of S. scandens was collected from Fuyang area of Zhejiang Province (30°05′2.4″N, 119°53′20.4″E) and deposited in the collection center of Zhejiang Chinese Medical University with the specific number of QLG-1919. Total genomic DNA was extracted and sequenced using the Illumina Hiseq Platform according to the previous report (Ying et al. Citation2019; Wang et al. Citation2020). The chloroplast genome of S. scandens was assembled by metaSPAdes with the chloroplast sequence of Dendrosenecio brassiciformis as reference (Nurk et al. Citation2017). The chloroplast was annotated using GeSqe and further confirmed by BLAST (Tillich et al. Citation2017). The complete cp genome of S. scandens was submitted to GenBank with the accession number of MT178410.
The length of the complete chloroplast genome sequence of S. scandens was 150,729 bp, with a large single-copy (LSC) region of 83,984 bp, a small single-copy (SSC) region of 17,835 bp, and two separated inverted repeated (IR) regions of 24,455 bp each. A total of 133 genes were identified in the cp of S. scandens, including 89 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The overall GC content was 37.4%, and the corresponding contents for LSC, SSC, and IR regions were 35.5%, 30.9% and 43%, respectively. The genome included 17 duplicated genes in the IR region and exhibited 52.48% protein-coding sequences. Moreover, a total of 38 small single repeats (SSR) are identified in the cp of S. scandens, ranging from 10 bp to 21 bp.
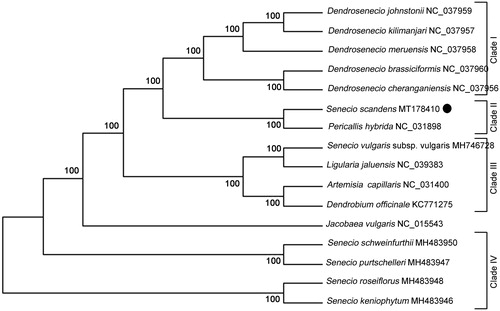
The complete genome sequences of S. scandens and other 14 representative species from the family Asteraceae were analyzed using MEGA 7.0 by maximum-likelihood (ML) method to confirm its phylogenetic position (Kumar et al. Citation2016). The result demonstrated that S. scandens clustered together with Pericallis hybrida and combined to form the clade II, indicating a close genetic relationship between the two species (). In addition, the six species from genus Senecio did not cluster together to form a monophyletic group, but was divided into three groups, providing molecular evidences for further revisions on genus Senecio (). Our results would contribute the development of molecular markers and further investigation on the population genetics and phylogenetics of the genus Senecio.
Figure 1. Phylogenetic tree position of the newly sequenced Senecio scandens analyzed by ML analysis of other representative species from family Asteraceae based on the complete chloroplast genomes. The species of Dendrobium officinale, Artemisia capillaris and Ligularia jaluensis were used as the out-group. Numbers on the nodes are bootstrap values from 100 replicates. The GenBank accession numbers are listed following the species name.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kumar S, Stecher G, Tamura K. 2016. Mega 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang D, Huang L, Chen S. 2013. Senecio scandens Buch.-Ham.: a review on its ethnopharmacology, phytochemistry, pharmacology, and toxicity. J Ethnopharmacol. 149(1):1–23.
- Wang Q, Yu S, Gao C, Ge Y, Cheng R. 2020. The complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Rubus chingii Hu. Mitochondrial DNA Part B. 5(2):1307–1308.
- Xiong A, Fang L, Yang X, Yang F, Qi M, Kang H, Yang L, Tsim KW, Wang Z. 2014. An application of target profiling analyses in the hepatotoxicity assessment of herbal medicines: comparative characteristic fingerprint and bile acid profiling of Senecio vulgaris L. and Senecio scandens Buch.-Ham. Anal Bioanal Chem. 406(29):7715–7727.
- Ying Z, Wang Q, Yu S, Liao G, Ge Y, Cheng R. 2019. The complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Lysimachia hemsleyana. Mitochondrial DNA B. 4(2):3878–3879.
