Abstract
The complete mitochondrial genome sequence of Microcotyle caudata was determined. The entire circular mitochondrial genome is 14,267 bp in length, comprising 12 protein-coding genes (PCGs, lacking atp8), 22 tRNA, 2 rRNA, several non-coding region, and one repeat sequence. The total length of the M. caudata mitochondrial genome was 140 bp shorter than a congeneric species, M. sebastis, but the arrangement of all 36 genes was identical with each other. The phylogenetic analysis of the concatenated 12 PCGs also revealed that M. caudata was most closely clustered with M. sebastis.
Microcotyle caudata (Monogenea: Microcotylidae) was first described as an ectoparasite of rockfish Sebastes sp. ( ), but later the host was re-described as dark-banded rockfish Sebastes inermis (Yamaguti Citation1934). Sebastes inermis is one of the commercially cultured fish species in Korea. In fact, a congeneric monogenean, Microcotyle sebastis is well known as a serious pathogen for Korean rockfish Sebastes schlegelii aquaculture in Korea (Kim and Cho Citation2000). Thus, M. caudata can also become a potential pathogen of S. inermis. In this study, we obtained the complete mitochondrial genome sequence of M. caudata.
The fish sample was collected from marine netpen cage located in Tongyeong, the southern coast of Korea (34.45 N, 128.24E) and M. caudata was collected from their gill lamellae. They were identified by morphological characters (Yamaguti Citation1934) and comparison with the mitochondrial DNA cox1 gene sequence registered in the public database (GenBank accession number: LC472527- LC472531). The slide specimen (specimen number IV00167588) was deposited in the Marine Biodiversity Institute of Korea. DNA sequencing was conducted with MGISEQ-2000 and the mitochondrial genome was assembled using Geneious v. 11.1.5 (Kearse et al. Citation2012). Annotation was performed by BLAST search against M. sebastis mitochondrial genome (GeneBank accession number: DQ412044), the MITOS (Bernt et al., Citation2013) and tRNAscan (Chan et al. Citation2019). The phylogenetic tree was constructed to elucidate the position of M. caudata on the basis of the concatenated amino acid sequences of protein-coding genes (PCGs) from 26 other mongenean mitochondrial genomes available in GenBank. Two Tricladida species (Crenobia alpine and Obama sp.) were used as outgroups.
The complete circular mitochondrial genome of M. caudata was 14,267 bp (GenBank accession number: MT180126), comprising 12 PCGs (lacking atp8), 22 tRNA genes, and 2 rRNA genes, several non-coding region and one repeat sequence. The total length of M. caudata mitochondrial genome was 140 bp shorter than that of M. sebastis, but the whole 36 gene arrangement of these 2 species were identical with each other: cox3-trnC-trnK-nad6-trnY-trnL1-trnS2-trnL2-trnR-nad5-trnE-cytb-nad4l-nad4-trnQ-trnF-atp6-nad2-trnV-trnA-trnD-nad1-trnN-trnP-trnI-nad3-trnS1-trnW-cox1-trnG-trnT-rrnL-rrnS-cox2-trnM-trnH. All the genes were transcribed from the same strand. The nucleotide composition was measured as 29.3% A, 41.5% T, 19.7% G and 9.5% C. The A + T content was calculated as 70.8%, similar with M. sebastis (70.5%, Park et al. Citation2007).
The total length of 12 PCGs was 10,287 bp, accounting for 72.1% of the entire mitochondrial genomic DNA. This is the same PCGs length of M. sebastis. The typical start codon ATG was found in 10 PCGs, whereas ATA found in nad4 and ATT found in cox1. Microcotyle caudata had 4 different stop codons: TAA (nad4l, nad4, atp6, nad2, nad3, cox1, cox3, nad6), TAG (cytb, cox2), TAT(nad1), and TTA(nad5). The size of 22 tRNA genes was 1,424 bp in total, ranging from 58 bp (trnS1) to 69 bp (trnL1). All tRNA genes had conventional secondary structure, but trnS1 lacked a DHU arm. The rrnL and rrnS were 976 bp and 721 bp in size, respectively. Several non-coding regions (520 bp in total) were also found and one repeat sequence (349 bp) was located between trnK (upstream) and nad6 (downstream).
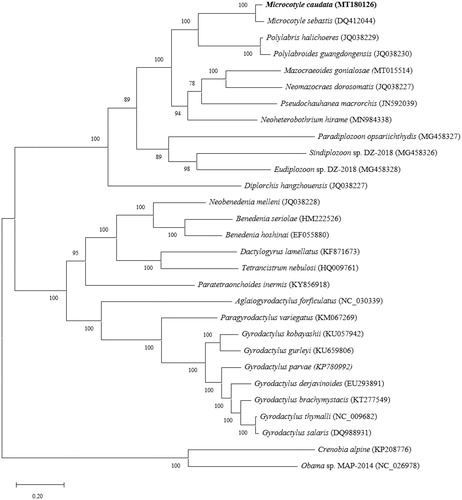
The phylogenetic analysis was conducted with the concatenated 12 PCGs sequences of M. caudata and other 26 monogeneans, by using MEGA X (Kumar et al. Citation2018). The results showed that M. caudata was most closely clustered with M. sebastis, forming a microcotylid subclade in the polyopisthocotylean clade ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chan PP, Lowe TMS. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kim KH, Cho JB. 2000. Treatment of Microcotyle sebastis (Monogenea: Polyopisthocotylea) infestation with praziquantel in an experimental cage simulating commercial rockfish Sebastes schlegeli culture conditions. Dis Aquat Org. 40:229–231.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Park JK, Kim KH, Kang S, Kim W, Eom KS, Littlewood D. 2007. A common origin of complex life cycles in parasitic flatworms: evidence from the complete mitochondrial genome of Microcotyle sebastis (Monogenea: Platyhelminthes). BMC Evol Biol. 7(1):11.
- Yamaguti S. 1934. Studies on the helminth fauna of Japan. Pt 2, Trematodes of fishes, I. Jap J Zool. 5:249–541.