Abstract
The complete mitochondrion DNA sequence of Lactarius lactarius was determined by next generation sequencing (NGS) platform as the first mitogenome report in the family. The mitogenome of L. lactarius wa 16,642 bp in length that contains 13 gene-encoded proteins, 2 ribosomal RNA gene (12S and 16S), 22 tRNA gene, and a control region (D-loop). Besides COX1 gene (GTG), all the other protein-coding genes showed typical (ATG) start codon. Incomplete stop condons (TA-/T–) were identified in COX2, COX3, ND2, ND3, ND4 and Cyt B. A phylogenetic analysis with currently reported mitogenomes of its relative species, L. lactarius formed a Lactariidae clade distinct from other families. Two species, Pentaceros japonicus (AB739063) and Banjos banjos (KT345965) were among the most closely related species with 86.61% and 86.33% sequence identity, respectively.
Lactarius lactarius (Bloch et al., Citation1801), commonly known as false travelly, is the sole member of the family Lactariidae based on the World Reister of Marine Species, WoRMS (http://www.marinespecies.org/index.php). The morphological characteristics of L. lactarius include the large head and mouth, compressed body, protruding lower jaw, forked caudal fin (Leis, Citation1994). This species is largely distributed from Indian Ocean to western Pacific Ocean which including Southeast Asian countries, Japan, China, and Australia. Since it is considered as commercially important fish resources in those countries, the scientific management of L. lactarius is important based on its genetic information.
The complete mitochondrial genome sequences L. lactarius was determined using high-thoughput sequencing (HTS) technique. The specimen was collected from the southern coastal water of the Korean peninsula (34°21′25.7″N 128°22′37.9″E) as part of a research survey funded by the Ministry of Oceans and Fisheries, Korea. Morphological similarity and cytochrome oxidase I (COI) sequence identity (JQ681391) confirmed identity of the species. The specimen and genomic DNA are stored at the Marine Biodiversity Institute of Korea (MABIK GR00004106). DNA was extracted from tissue by DNA isolation kit (Abcam, UK) and sheared by Covaris M220 Focused-Ultrasonicator (Covaris Inc., San Diego, CA). TruSeq® RNA library preparation kit V2 for library preparation and MiSeq platform (Illumina, San Diego, CA) was employed for sequencing. Bioinformatics software, Geneious® 11.0.2 (Kearse et al., Citation2012)was used for mapping, analyzing and final assembly of the sequence. The tRNAScan-SE software (Lowe and Chan, Citation2016) predicted structural details of 22 tRNAs. A phylogenetic tree was built with minimum evolution (ME) algorithm using MEGA7.0 programs (Kumar et al., Citation2016).
The complete mitochondrial genome of L. lactarius (MN952976) was 16,642 bp in length encoding 37 eukaryotic-canonical genes including 13 proteins (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs). Twenty-eight genes were located on Heavy (H) strand, while 9 were on the light (L) strand. Among 13 PCGs (11,439bp), unusual start codon (GTG) was identified only in COX1 gene. Incomplete stop condons (TA-/T–) were identified in COX2, COX3, ND2, ND3, ND4 and Cyt B. All the tRNAs have a clover-leaf molecular structure excluding the tRNASer-GCT. It has a 965 bp long D-loop (control region) with nucleotide composition of A:31.8%,T:30.5%, G: 16.3%, C:21.5%. A 33 bp of light strand replication origins (OL) was located within WANCY tRNAs cluster.
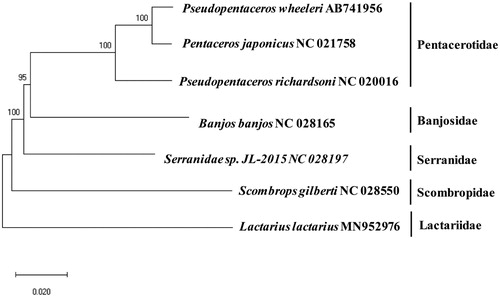
A phylogenetic tree constructed with its relative mitogenome sequences displayed L. lactarius as a distict clade from other families (). Although this is a sole member of the Lactariidae family, the addition mitogenome sequences should be supplemented to extend our knowledge about its genetic structures considering its wide distribution from Indian to Pacific Oceans. Besides the family, Pentaceros japonicus (AB739063) and Banjos banjos (KT345965) with 86.61% and 86.33% sequence identity, respectively.
Figure 1. Phylogenetic tree of Lactarius lactarius with its relative species: Complete mitogenomes were retrieved from GenBank to construct a phylogenetic tree by using MEGA7 software using Minimum Evolution (ME) algorithm with 1,000 bootstrap replications. All mtDNA sequence of species has its GenBank accession numbers with the scientific name.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bloch ME, Hennig JF, Schneider JG. 1801. M.E. Blochii … Systema ichthyologiae iconibus CX illustratum. Berolini: Sumtibus auctoris impressum et Bibliopolio Sanderiano commissum.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England). 28(12):1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Leis JM. 1994. Larvae, adults and relationships of the monotypic perciform fish family Lactariidae. Rec Aust Mus. 46(2):131–143.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
