Abstract
The mitogenome was sequenced through whole-genome shotgun sequencing using the Illumina platform, and phylogenetic analysis was carried out on the basis of 13 PCGs and two rRNAs. The complete T. cardis mitogenome comprised 16,761 bp, MN865118. The organization and location of genes in the T. cardis mitogenome were consistent with those reported for other Turdidae species. Phylogenetic relationships based on Bayesian inference and the Maximum likelihood method revealed that T. cardis was closely related to Turdus hortulorum, indicating that the molecular taxonomy of T. cardis is consistent with its current morphological characteristics. Our mitogenomic data may have applications in further phylogenetic and taxonomic studies of Turdidae.
Keywords:
The Grey thrush (Turdus cardis) belongs to the order Passeriformes and family Turdidae. It is distributed in Japan, Russia, North Korea, South Korea, Thailand, Vietnam, and Laos and in the central, eastern, and southern regions of China. Most of these birds dwell in shrubs and forests at altitudes of 500–800 m. The Grey thrush is reportedly timid, easily frightened, and has a pleasant song. They are generally solitary; however, they form small groups during migration. They primarily feed on insects, thus helping to control regional pests and facilitating the integrated management of forest diseases and insect pests (Miyazawa Citation1971).
In this study, the complete mitogenome sequence of T. cardis was determined. The specimens were collected from Nanjing Zhongshan Botanical Garden, Jiangsu province, China, on November 1, 2019 (32°3′20.95″N latitude and 118°49′37.45″E longitude. After sampling, the specimens (NJFU-2020-WHD) were stored in the Animal Specimens Museum of Nanjing Forestry University.
The complete mitogenome of T. cardis contained 16,761 base pairs (bp) and was submitted to GenBank (accession number MN865118). The T. cardis mitogenome was circular and contained 37 mitochondrial genes (13 PCGs, 22 tRNA genes, and 2 rRNA genes) and 1 major noncoding region (D-loop). The position of each gene in the mitogenome was identical to that in other Turdidae species. One of the 13 PCGs (nad6) and 8 tRNAs (trnA, trnC, trnE, trnN, trnP, trnQ, trnS2, and trnY) were encoded on the L-strand, whereas the other 28 genes, including 12 PCGs, 14 tRNAs, 2 rRNAs, and 1 D-loop, were encoded on the H-strand. The nucleotide composition of the complete mitogenome was as follows: A = 4905 (29.26%), T = 3909 (23.32%), G = 2496 (14.89%), C = 5451 (32.52%). The entire mitogenome had a high A + T content of 52.59%, including 51.52% in PCGs, 57.05% in tRNA genes, 54.02% in rRNA genes, and 54.65% in the D-loop, concurrent with the typical base bias of avian mitogenomes (Haring et al. Citation2001; Bi et al. Citation2019; Liu et al. Citation2019; Sun et al. Citation2019a, 2019b).
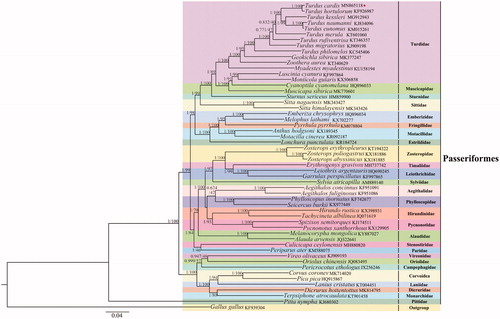
To elucidate the phylogenetic inter-relationships within the orders Turdidae and Passeriformes, we obtained the concatenated nucleotide sequences of 13 PCGs and 2 rRNAs from 52 Passeriformes species. Additionally, we used G. gallus as an outgroup because it is distant from the classification of orders Galliformes and Passeriformes (Reyes et al. Citation2005; Kan et al. Citation2010). Both BI and ML analyses revealed identical topologies (). Target species T. cardis and T. hortulorum were clustered into one branch with high nodal support value (BI posterior probabilities = 1; ML bootstrap = 100). Moreover, the (T. cardis + T. hortulorum) clade and (T. kessleri + [T. naumanni + T. eunomus]) clade displayed a sister-group relationship. T. merula and the clade ([T. cardis + T. hortulorum] + [T. kessleri + (T. naumanni + T. eunomus)]) clustered in one branch. T. rufiventris clustered in one branch with the clade (T. merula + [(T. cardis + T. hortulorum) + (T. kessleri + [T. naumanni + T. eunomus])]). In the branch (T. philomelos + [T. migratorius + (T. rufiventris + [T. merula + ([T. cardis + T. hortulorum] + [T. kessleri + (T. naumanni + T. eunomus)])])]), the aforementioned nine species belonged to the genus Turdus. Furthermore, T. philomelos displayed close relationships with Zoothera aurea, Geokichla sibirica, and Myadestes myadestinus, indicating that the four species belonged to the family Turdidae. Our results provide insights into the T. cardis mitogenome and evolutionary relationships within the family Turdidae, enabling further evolutionary studies of Passeriformes and Turdidae.
Figure 1. Phylogenetic trees of Passeriformes species using concatenated nucleotide sequences of 13 protein-coding genes and 2 rRNAs. Numbers beside the nodes are posterior probabilities (Bayesian interference, BI) and bootstrap (maximum likelihood, ML). GenBank accession numbers of mitogenomes used are.
Disclosure statement
The authors declare that they have no competing interests. The sequence has been submitted to NCBI under the accession no. MN865118.
Additional information
Funding
References
- Bi D, Ding H, Wang Q, Jiang L, Lu W, Wu X, Zhu R, Zeng J, Zhou S, Yang X, et al. 2019. Two new mitogenomes of Picidae (Aves, Piciformes): sequence, structure and phylogenetic analyses. Int J Biol Macromol. 133:683–692.
- Haring E, Kruckenhauser L, Gamauf A, Riesing MJ, Pinsker W. 2001. The complete sequence of the mitochondrial genome of Buteo buteo (Aves, Accipitridae) indicates an early split in the phylogeny of raptors. Mol Biol Evol. 18(10):1892–1904.
- Kan XZ, Li XF, Zhang LQ, Chen L, Qian CJ, Zhang XW, Wang L. 2010. Characterization of the complete mitochondrial genome of the Rock pigeon, Columba livia (Columbiformes: Columbidae). Genet Mol Res. 9(2):1234–1249.
- Liu H, Jin K, Li L. 2019. The complete mitochondrial genome of the Fischer’s Lovebird Agapornis fischeri (Psittaciformes: Psittacidae). Mitochondrial DNA Part B. 4(1):1217–1218.
- Miyazawa K. 1971. Life history of the Grey Thrush Turdus cardis. J Yamashina Inst Ornithol. 6(3):300–315.
- Reyes A, Yang MY, Bowmaker M, Holt IJ. 2005. Bidirectional replication initiates at sites throughout the mitochondrial genome of birds. J Biol Chem. 280(5):3242–3250.
- Sun CH, Liu B, Lu CH. 2019a. Complete mitochondrial genome of the Siberian thrush, Geokichla sibirica sibirica (Aves, Turdidae). Mitochondrial DNA Part B. 4(1):1150–1151.
- Sun CH, Liu HY, Lu CH. 2019b. Five new mitogenomes of Phylloscopus (Passeriformes, Phylloscopidae): Sequence, structure, and phylogenetic analyses. Int J Biol Macromol. 146(1):638–647.
