Abstract
Siniperca fortis (Perciformes: Sinipercidae) is an endemic species in Zhujiang (Pearl) River system of China. Through genome sequence and phylogenetic analyses, the main structural characteristics of the mitochondrial genome sequence and the phylogenetic information of S. fortis were revealed. In this study, the complete mitochondrial genome of S. fortis was firstly determined. The genome is 16,495 bp in length and consists of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 1 control region. The genetic distance and the ML Phylogenetic tree of 11 species in Sinipercidae supported that S. fortis was a valid species. The mitochondrial DNA information would be useful in species identification and natural resources conservation.
Keywords:
Sinipercids are an endemic small group of 12 freshwater perciform fish species in East Asia that have attracted much attention from taxonomists (Zhao et al. Citation2008; Chen et al. Citation2012). These species were once assigned to only one genus, Siniperca (Zheng Citation1989), or to two genera Siniperca, Coreoperca (Chen et al. Citation2007), or even three genera, Siniperca, Coreoperca, Coreosiniperca (Kong and Zhou, Citation1993). Siniperca fortis, as one of the main species of Sinipercids, is a species endemic to the Zhujiang (Pearl) River system in China. Siniperca fortis is distributesdonly in Duliujiang and Rongjiang (Dai Citation2011). There has been debate about the validity of S. fortis. Some researchers classified S. fortis as S. obscura (Zhou et al. Citation1988), and some researchers considered S. fortis a valid species (Liu and Chen Citation1994). There are a few studies on the germplasm resources of S. fortis (Chen et al. Citation2010), but no report on mitochondrial genes of Siniperca fortis is available.
In this study, the complete mitochondrial DNA sequence of S. fortis was firstly determined. The sample was collected from Rongjiang system, Guangxi province, China (25°51′35.59″N, 109°42′38.17″E). Muscle samples (#L2015100002) were fixed in 95% ethanol and preserved in Fisheries Ecological Environment Laboratory, Guangxi Academy of Fisheries Sciences. The complete mitochondrial genomes of S. fortis were amplified and sequenced using 13 primer pairs. The complete mtDNA sequence of S. fortis was 16,495 bp in length and has been deposited in GenBank (accession number: MN857459), which contained 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes (12S and 16S rRNA), and 1 control region. The overall base composition of mtDNA included T (25.81%), C (29.32%), A (28.43%), and G (16.44%), with the percentage of G and C (45.76%) being slightly lower than that of A and T (54.24%).
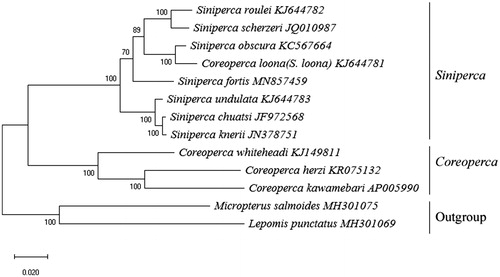
Based on the mitochondrion of 11 Sinipercidae species, we analyzed the genetic distance and clustering using Mega X soft (). Sinipercidae with two genera, Siniperca and Coreoperca was supported by the ML Phylogenetic tree of 11 species in Sinipercidae. Siniperca was consisted of two subclades: a subclade having S. undulata sister to S. chuatsi and S. kneri. Within another subclade containing the other members of the genus, S. roulei and S. scherzeri sister to S. obscura and C.loona (=S.loona), and finally next joined by S. fortis. By comparing the genetic distance between the S. fortis and other species, all genetic distance between S. fortis and 10 other species were over 0.05, we consider that S. fortis is a valid species, rather than a synonym of S. obscura. The complete mitochondrial genome sequence of S. fortis will be useful for its genetic research and future conservation and management.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chen D, Guo X, Nie P. 2007. Non-monophyly of fish in the Sinipercidae (Perciformes) as inferred from cytochrome b gene. Hydrobiologia. 583(1):77–89.
- Chen D, Guo X, Nie P. 2010. Phylogenetic studies of sinipercid fish (Perciformes: Sinipercidae) based on multiple genes, with first application of an immune-related gene, the virus-induced protein (viperin) gene. Mol Phylogenet Evol. 55(3):167–1176.
- Chen D-X, Chu W-Y, Liu X-L, Nong X-X, Li Y-L, Du S-J, Zhang J-S. 2012. Phylogenetic studies of three sinipercid fishes (Perciformes: Sinipercidae) based on complete mitochondrial DNA sequences. Mitochondrial DNA. 23(2):70–76.
- Dai Y. 2011. Fish fauna and zoogeographical division in the DuliuJ, Guizhou, China. J Fish China. 35(6):871–879. (In Chinese).
- Kong X, Zhou C. 1993. Comparative studies on the skeletal characteristics of seven Sinipercinae fish of China. J Ocean Univ Qingdao. 23(3):116–124. (In Chinese with English abstract).
- Liu H, Chen Y. 1994. Phylogeny of the sinipercine fish with some taxonomic notes. Zool Res. 15(Suppl):1–12.
- Zhao JH, Li CH, Zhao LL, Wang WW, Cao Y. 2008. Mitochondrial diversity and phylogeography of the Chinese perch, Siniperca chuatsi (Perciformes: Sinipercidae). Mol Phylogenet Evol. 49(1):399–404.
- Zheng C. 1989. Fish of the Zhujiang River. Beijing: Science Press. (In Chinese).
- Zhou CW, Yang Q, Cai D. 1988. On the classification and distribution of the Sinipercinae fishes (family serranidae). Zool Res. 9(2):113–125. (Chinese).