Abstract
Styrax japonicus (Styraceae) is a deciduous tree that distributed in East Asia, and grown at altitudes between 300 and 1800 meter in China. In this study, the complete chloroplast genome of S. japonicus is reported. The whole genome with size is 157,929 bp contains a pair of inverted repeats (IRs) of 26,054 bp, separated by a large single copy region (LSC) of 87,540 bp and a small single copy region (SSC) of 18,281 bp. The genome contains 131 genes, including 85 protein-coding genes, 38 transfer RNA genes, and 8 ribosomal RNA genes. Phylogenetic analysis supported that the S. japonicus closely clustered with S. grandifloras, S. confuses and S. calvescens.
Styrax japonicus Sieb. et Zucc. (Styraceae) is a deciduous tree that is widely distributed in southern area of the Yellow River in China with the altitude ranged from 300 m to 1800 m. In consideration of the fragrant and beautiful flowers, it is a potential ornamental tree and has a good application prospect in the garden construction (Cui and Zhao Citation1998; Fritsch et al. Citation2001). Previous studies have conducted the phylogenetic and biogeographical analyses for S. japonicus and its closely-related species by using isozymes and internal transcribed spacer region (ITS) (Fritsch Citation1996, Citation2001), and developed many microsatellite loci (Wang et al. Citation2010). However, little genetic information, especially from the genome, is available so far. Here, we reported the complete chloroplast genome of S. japonicus (the GenBank accession number is MT178456), which will contribute to the molecular and phylogenetic study in the future.
Fresh leaves of S. japonicus were collected from Nanyue county, Hunan province, China (coordinates: 27°16′4.66″ N, 112°43′1.05″ E), and the voucher specimens were deposited in the Herbarium of Central South University of Forestry and Technology (accession number is YML-20190812). Total genomic DNA was isolated using modified CTAB method (Doyle and Doyle Citation1987), and the paired end (PE) DNA library was constructed and sequenced on the Illumina Hiseq Platform (Novogene, Beijing, China). The published chloroplast genomes of Styrax suberifolius (GenBank accession number is MG719828) was detected as the best reference, and further used for assembled via NOVOPlasty program (Dierckxsens et al. Citation2017). The software Geneious v11.0.3 (Kearse et al. Citation2012) was employed to annotate the assembled plastid genome. After careful inspection and manual correction, the genome map was generated using the web server OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013).
The whole chloroplast genome of S. japonicus was 157,929 bp in size, consisting of a pair of inverted repeat regions (IRs, 26,054 bp), which were separated by a large single copy (LSC, 87,540 bp) and a small single copy (SSC, 18,281 bp). A total of 131 genes were detected, including 85 protein-coding genes, 38 transfer RNA genes, and 8 ribosomal RNA genes. Most of the genes exhibit as a single copy in LSC or SSC region, However, 17 genes are duplicated, including 6 protein-coding genes, 4 ribosomal RNA genes and 7 transfer RNA genes. GC content of the complete genome was 37.0%, which in LSC, SSC and IR regions were 34.8%, 30.3% and 42.9%, respectively.
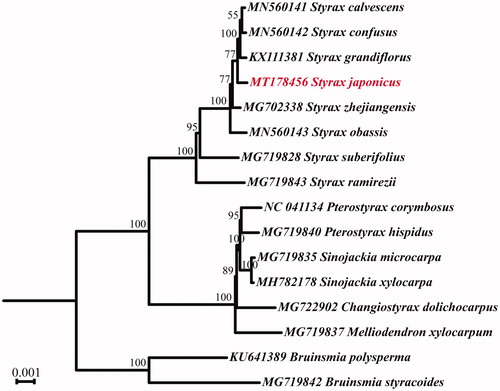
In order to clarify the phylogenetic relationship between S. japonicus and other Styracaceae species, 15 cp genome sequences were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/). After aligned using MAFFT (Katoh et al. Citation2002), the alignment was used to generate a maximum likelihood (ML) tree via RAxML v8 (Stamatakis Citation2014) with 1000 bootstrap replicates. (). The result revealed that S. japonicus was closely cluster with S. grandifloras, S. confuses and S. calvescens. The cp genome of S. japonicus will provide fundamental information for the phylogenetic and evolution studies of Styracaceae.
Disclosure statement
The authors are grateful to the opened raw genome data from public database. The authors declare no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Cui JR, Zhao SW. 1998. The introduction and cultivation of the Styrax japonicus. Beijing Garden. 3:13.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18–e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fritsch P. 1996. Isozyme analysis of intercontinental disjuncts within Styrax (Styracaceae): implications for the Madrean-Tethyan hypothesis. Am J Bot. 83(3):342–355.
- Fritsch PW, Morton CM, Chen T, Meldrum C. 2001. Phylogeny and biogeography of the Styracaceae. Int J Plant Sci. 162(S6):S95–S116.
- Fritsch PW. 2001. Phylogeny and biogeography of the flowering plant genus Styrax (Styracaceae) based on chloroplast DNA restriction sites and DNA sequences of the internal transcribed spacer region. Mol Phylogenet Evol. 19(3):387–408.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang XY, Yu S, Liu M, Yang QS, Chen XY. 2010. Twenty-three microsatellite loci for Styrax confuses and Styrax japonicus (Styracaceae). Conservation Genet Resour. 2(S1):51–54.