Abstract
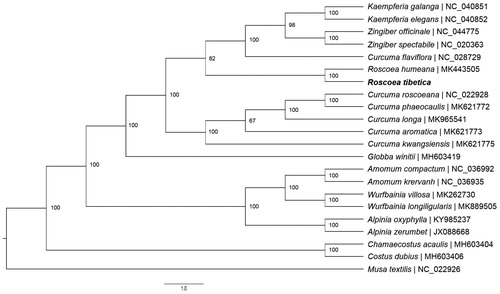
Roscoea tibetica is a most widespread and extensively phenotypic-variable alpine ginger species. Here, we reported the complete chloroplast (cp) genome sequence of R. tibetica. The complete chloroplast genome is 163,515 base pairs (bp) in length, containing two inverted repeat (IRa and IRb) regions of 29,781 bp, which was separated by a large single-copy (LSC) region of 88,090 bp and a small single-copy (SSC) region of 15,863 bp. The overall GC content of the plastid genome was 36.06%. In total, 113 unique genes were annotated, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Phylogenetic analysis based on 22 chloroplast genomes indicates that R. tibetiaca is closely related to R. humeana, forming a monophyletic group with high value.
The ginger family (Zingiberaceae) is largely restricted to the tropical lowlands of Asia, Africa, and Americas. The Roscoea (∼20 species) is a unique genus in ginger family that occurs in subtropical alpine regions (Kress et al. Citation2002). The genus is an ideal study system for investigating a wide range of questions relating to the evolution and ecology because of its evolutionary history, diversity floral traits and disjunct distribution (Zhang and Li Citation2008; Zhang et al. Citation2011; Zhao et al Citation2016; Paudel et al. Citation2018; Li et al. Citation2020). However, very little genetic and genomic information has hindered the studies in the genus. Roscoea tibetica is a most widespread and extensively phenotypic-variable species in the genus. Here, we reported a complete plastome of R. tibetica and revealed its phylogenetic relationships with related species.
We collected fresh leaves of R. tibetica from Xuesong Village, Lijiang, Yunnan, China (27.011°N, 100.222°E). Voucher specimen was deposited at the herbarium of Yunnan Key Laboratory of Plant Reproductive Adaption and Evolutionary Ecology, Yunnan University (specimen code: R84-2016). Genomic DNA was extracted from the fresh leaves using the modified CTAB method (Doyle and Doyle Citation1987). Subsequently, the cp genome was amplified using long-range PCR with 15 runs. Paired-end sequencing (250 bp) was performed on the Illumina MiSeq 2000. High-quality reads were assembled into contigs using the de novo assembler in CLC Genomics Workbench v6.5 (CLC Bio), using a k-mer of 64 and a minimum contig length of 500 bp. The de novo contigs were assembled into complete chloroplast genomes following the procedure of Yang et al. (Citation2004). The complete cp genome was annotated using the program DOGMA (Wyman et al. Citation2004).
The complete cp genome of R. tibetica composed of single circular double-stranded DNA molecules and was 163,515 bp in length. The overall GC content of the plastid genome was 36.1%. The data are deposited in GenBank with the accession number MT180128. Similar to R. humeana (Zhu et al. Citation2019), the complete chloroplast contains two inverted repeat (IRa and IRb) regions of 29,781 bp, which was separated by and a large single-copy (LSC) region of 88,090 bp and a small single-copy (SSC) region of 15,863 bp. There was a total of 113 unique genes, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes, in R. tibetica cp genome. Among these genes, ten protein-coding genes and seven tRNA genes contained a single intron, and three protein-coding genes including rps12, ycf3 and clpP possessed two introns. Moreover, the ycf1, located in the range of 113,954–117,847 bp was identified as pseudogenes because of the partial duplication. We used RAxML (Stamatakis Citation2014) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of the chloroplast sequence of R. tibetica and 19 published cp genomes from Zingiberaceae (17 species) and Costaceae (2 species), using one species in Musaceae (Musa textilis) as outgroups. The ML tree indicated that R. tibetiaca is closely related to R. humeana, forming a monophyletic group with high value (). The availability of complete plastome sequence of R. tibetica will be helpful for further studies on ecology and evolution of alpine gingers.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Kress WJ, Prince LM, Williams KJ. 2002. The phylogeny and a new classification of the gingers (Zingiberaceae): evidence from molecular data. Am J Bot. 89(10):1682–1696.
- Li D‐B, Ou X‐K, Zhao J‐L, Li Q‐J. 2020. An ecological barrier between the Himalayas and the Hengduan Mountains maintains the disjunct distribution of Roscoea. J Biogeogr. 47(2):326–341.
- Paudel BR, Burd M, Shrestha M, Dyer AG, Li Q‐J. 2018. Reproductive isolation in alpine gingers: How do coexisting Roscoea (R. purpurea and R. tumjensis) conserve species integrity?. Evolution. 72(9):1840–1850.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yang SX, Yang JB, Lei LG, Li DZ, Yoshino H, Ikeda T. 2004. Reassessing the relationships between Gordonia and Polyspora (Theaceae) based on the combined analyses of molecular data from the nuclear, plastid and mitochondrial genomes. Plant Syst E. 248:45–55.
- Zhang Z-Q, Kress WJ, Xie W-J, Ren P-Y, Gao J-Y, Li Q-J. 2011. Reproductive biology of two Himalayan alpine gingers (Roscoea spp., Zingiberaceae) in China: pollination syndrome and compensatory floral mechanisms. Plant Biol (Stuttg). 13(4):582–589.
- Zhang Z-Q, Li Q-J. 2008. Autonomous selfing provides reproductive assurance in an alpine ginger Roscoea schneideriana (Zingiberaceae). Ann Bot. 102(4):531–538.
- Zhao J-L, Xia Y-M, Cannon CH, Kress WJ, Li Q-J. 2016. Evolutionary diversification of alpine ginger reflects the early uplift of the Himalayan–Tibetan plateau and rapid extrusion of Indochina. Gondwana Res. 32:232–241.
- Zhu X-F, Yu X-Q, Zhao J-L. 2019. The complete chloroplast sequence of Roscoea humeana (Zingiberaceae): an alpine ginger in the Hengduan Mountains. China. Mitochondrial DNA Part B. 4(1):1398–1399.