Abstract
Viburnum odoratissinum has very high ornamental and medicinal value. In this study, the chloroplast (cp) genome sequence of V. odoratissinum was characterized using Illumina pair-end sequencing. The complete cp genome was 158,757 bp in length, containing a large single-copy region (LSC) of 87,350 bp and a small single-copy region (SSC) of 18,409 bp, which were separated by a pair of 26,499 bp inverted repeat regions (IRs). The overall GC content was 38.10%. The genome contained 132 genes, including 87 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Further, phylogenetic analysis suggested that the V. odoratissinum has a close relationship with Viburnum betulifolium, which belong to Adoxaceae.
The Viburnum odoratissimum, belonging to the genus Viburnum, is an evergreen shrub or dungarunga mainly distributed in Southeast of China (Ma et al. Citation2014), which has been widely cultivated as excellent landscape plant in China. In addition, V. odoratissimum can be used as folk medicine for their biological activities, such as anti-cancer, anti-inflammatory, antibacterial, antinociceptive and neuroprotective and others (Y. Zhang et al. Citation2018; Y.Y. Zhang et al. Citation2019). However, the phylogenetic position of V. odoratissimum is still unresolved. V. odoratissimum was originally calssified as the Caprifoliaceae family, but it has been divided into the family Adoxaceae based on molecular-level taxonomic research in recent years (Winkworth and Donoghue Citation2005). Therefore, we first reported the complete chloroplast genome (cp) of V. odoratissimum and reveal its phylogenetic relationship with related species, which would be helpful for its evolution and biological research.
The fresh leaves of V. odoratissimum were collected from City Forest Park in Fuyang, in Hangzhou city, Zhejiang province of China (119°58′8.4″E, 30°4′20.28″N). The Voucher specimen and DNA sample were stored in the Herbarium of College of pharmaceutical sciences, Zhejiang Chinese Medical University (specimen code ZCMU4C505). Genomic DNA was isolated by the modified method CTAB (Doyle and Doyle Citation1987). Sequencing was performed with an Illumina Hiseq 4000 platform (Biodata Biotechnologies Inc., Hefei, China), yielding at least 6.79 GB of clean data for V. odoratissimum. The filtered reads were assembled using the program NOVOPlasty (Dierckxsens et al. Citation2017) with complete chloroplast genome of its close relative Viburnum betulifolium as the reference (GenBank accession number NC_037951.1). CpGAVAS was used to annotate the sequences (Chang et al. Citation2012); DOGMA and BLAST were used to check the results of the annotation (Wyman et al. Citation2004). tRNAscanSE was used to identify the tRNAs (Schattner et al. Citation2005).
The size of the complete chloroplast genome of V. odoratissimum (GenBank accession number MN894600) was 158,757 bp. The cp genome displayed a typical quadripartite structure, including a pair of IR (26,499 bp) separated by the large single-copy (LSC; 87,350 bp) and small single-copy (SSC; 18,409 bp) regions. The overall GC content of chloroplast genome is 38.10%, and while it is 42.98% of IRs, which was higher than LSC and SSC regions (36.41 and 32.12%, respectively). In the V. odoratissimum chloroplast genome, 132 functional genes were observed, including 8 rRNA genes, 37 tRNA genes, and 87 protein-coding genes, of which 18 were duplicated in the IRs region. Among these genes, 19 genes contain a single intron and 4 genes contain two introns.
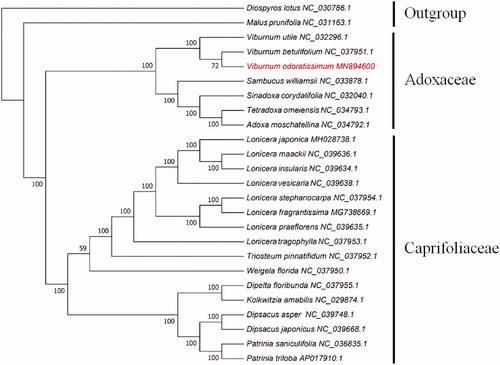
To ascertain the phylogenetic position of V. odoratissimum, we conducted a phylogenetic analysis with the maximum likelihood methods based on the alignments created by the MAFFT (Katoh et al. Citation2005) within the 25 complete chloroplast genomes from the NCBI Organelle Genome and Nucleotide Resources database (). NJ phylogenetic tree was performed using MEGA7 (Kumar et al. Citation2016) with 1000 bootstrap replicates. The results indicated that V. odoratissimum has a close relationship with Viburnum betulifolium, which are all belong to Adoxaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chang L, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13(1):715.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger dataset. Mol Biol Evol. 33(7):1870–1874.
- Ma J, Yang X, Zhang J, Liu X, Deng L, Shen X, Xu G. 2014. Sterols and terpenoids from Viburnum odoratissimum. Nat Prod Bioprospect. 4(3):175–180.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–9.
- Winkworth RC, Donoghue MJ. 2005. Viburnum phylogeny based on combined molecular data: implications for taxonomy and biogeography. Am J Bot. 92(4):653–666.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhang Y, Zhou WY, Song XY, Yao GD, Song SJ. 2018. Neuroprotective terpenoids from the leaves of Viburnum odoratissimum. Nat Prod Res. 10:1–8.
- Zhang YY, Chen JJ, Li DQ, Zhang Y, Wang XB, Yao GD, Song SJ. 2019. Network pharmacology uncovers anti-cancer activity of vibsane-type diterpenes from Viburnum odoratissimum. Nat Prod Res. 11:1–4.