Abstract
Corybas taliensis is an endangered terrestrial orchid, endemic to China. In this study, we assembled the complete chloroplast (cp) genome using Illumina pair-end sequencing data. The whole cp genome is 131,393 bp in length with 38.21% of GC content. It encodes 121 genes, including 38 tRNA genes, 8 rRNA genes, and 75 protein-coding genes. The phylogenetic tree based on 13 cp genomes of Orchidoideae suggested that C. taliensis is closely related to Rhizanthella gardneri. This study will pave the way to understand the genomic information and conservation of this endangered endemic orchid species.
Corybas is one of the most extraordinary genus of Orchidaceae. These terrestrial orchids group consist of a single leaf and flower. Corybas comprises about 160 species mainly distributed in Australia, New Guinea and the Pacific islands, extending through southeast of Asia to the Himalayas (Lehnebach et al. Citation2016). In China, Corybas represented with five species, including an endemic Corybas taliensis T. Tang (Chen et al. Citation2009). C. taliensis is small herbaceous orchid with exceptionally beautiful fascinating colorful sepal and the heart-shaped leaf with white veins (Tang and Wang Citation1951). This orchid always grows among mosses, which is sensitive to the changing environment, resulting in its extremely limited distribution range and high extinction risk (Go et al. Citation2015). This species has been categorized an Endangered (EN) in Threatened Species List of China’s Higher Plants following the IUCN Red List categories and criteria (Qin et al. Citation2017). C. taliensis also was listed as a PSESP (plant species with extremely small populations) in the China’s National Conservation Program (2017FY100100), for an emergency rescue plan of the threatened species (Sun et al. Citation2019). However, we have not seen any research or conservation work about this endemic orchid species so far. In this study, we reported the complete chloroplast genome of C. taliensis. These genomic resources will contribute for the conservation of this endangered species.
The fresh leaf of wild C. taliensis was collected from Gaoligong Mountain, Tengchong, Yunnan Province of China (98°42′E, 25°46′N), and dried using silica gel. The voucher specimen (KIBDZL18Z01) was deposited in Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming. Genomic DNA was extracted from ∼4 mg dried leaf using Plant Genomic DNA Kit (Tiangen Biotech, Beijing). DNA sequencing was performed on the Illumina Hiseq 2500 platform (Illumina, San Diego, USA). The screened sequences used for de novo assembly to reconstruct the cp genome using SPAdes v3.6.1 (Bankevich et al. Citation2012). Automatic annotation was generated by CpGAVAS (Liu et al. Citation2012). The potential microsatellites identified using MISA software (http://pgrc.ipk-gatersleben.de/misa/).
The cp genome sequence of C. taliensis is 131,393 bp in length, and the total GC content is 38.21%. The cp genome encodes 121 genes, including 38 tRNA genes, 8 rRNA genes, and 75 protein-coding genes. Most genes occur in a single copy, while 18 genes are duplicated and rps15 is presented in three copies. 60 SSR markers were found in the C. taliensis cp genome. The complete cp genome was deposited in GenBank with the accession number MN783368.
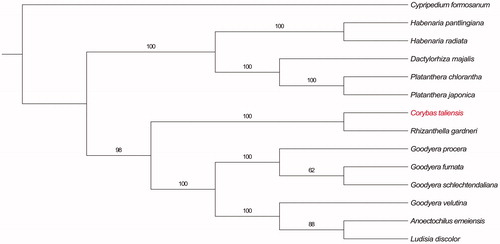
To reveal the phylogenetic position of C. taliensis, we chose 12 published cp genomes from subfamily Orchidoideae, with Cypripedium formosanum from subfamily Cypripedioideae as outgroup. The maximum likelihood (ML) tree with 1000 rapid bootstrap replications was performed with RAxML v8.2.10 in CIPRES Science Gateway (Stamatakis Citation2014). The results showed that C. taliensis is closely related to Rhizanthella gardneri (), an underground orchid with rampant cp gene loss (Delannoy et al. Citation2011).
Figure 1. The ML tree based on 13 cp genomes of Orchidoideae. Numbers at nodes indicate bootstrap values. Accession number: Cypripedium formosanum (KJ501998), Platanthera chlorantha (MK937914), Platanthera japonica (MG925368), Dactylorhiza majalis (MK984209), Habenaria pantlingiana (KJ524104), Habenaria radiata (KX871237), Corybas taliensis (MN783368), Rhizanthella gardneri (GQ413967), Goodyera procera (KT886429), Goodyera fumata (KJ501999), Goodyera schlechtendaliana (KT886431), Goodyera velutina (KT886432), Anoectochilus emeiensis (LC057212), Ludisia discolor (KU578274).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comp Biol. 19(5):455–477.
- Chen SQ, Gale SW, Cribb PJ. 2009. Flora of China, vol. 25. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press; p. 86–87.
- Delannoy E, Fujii S, Colas Des Francs-Small C, Brundrett M, Small I. 2011. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 28(7):2077–2086.
- Go R, Ching TM, Nuruddin AA, Abdullah JO, Jin NY, Nordin FA, Eng KH, Nulit R. 2015. Extinction risks and conservation status of Corybas (Orchidaceae; Orchidoideae; Diurideae) in Peninsular Malaysia. Phytotaxa. 233(3):273–280.
- Lehnebach CA, Zeller AJ, Frericks J, Ritchie P. 2016. Five new species of Corybas (Diurideae, Orchidaceae) endemic to New Zealand and phylogeny of the Nematoceras clade. Phytotaxa. 270(1):1–24.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Qin H, Yang Y, Dong S, He Q, Jia Y, Zhao L, Yu S, Liu H, Liu B, Yan Y, et al. 2017. Threatened species list of China’s higher plants. Biodivers Sci. 25(7):696–744.
- Stamatakis A. 2014. Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sun WB, Yang J, Dao ZL. 2019. Study and conservation of plant species with extremely small population (PSESP) in Yunnan province, China. Beijing (China): Science Press; p. 143.
- Tang T, Wang FT. 1951. Corybas Salisb., a new addition to the orchid flora of China. Acta Phytotaxon Sin. 1:185–187.
