Abstract
The complete chloroplast genome (plastome) of a deciduous shrub Lespedeza bicolor (commonly called bush clover) was determined in this study. The plastome was 148,930 bp in size with a canonical quadripartite structure, including two inverted repeats regions of 23,827 bp for each, a large single-copy region of 82,338 bp in length, and a small single-copy region of 18,938 bp in length. The overall guanine-cytosine (GC) content of this plastome was 35.0%. A total of 112 unique genes was annotated in this plastome, including four rRNA genes, 30 tRNA genes, and 78 protein-coding genes. Among them, two protein-coding genes (clpP and ycf3) contained two introns, and eight protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, and rps16) and six tRNA genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained one intron. ML phylogenetic analysis showed that L. bicolor was sister to other Lespedeza species.
Keywords:
The genus Lespedeza, belonging to Fabaceae family, contained ca. 40 species with high economic and medicinal value (Xu, et al. Citation2012). Species in this genus are mainly distributed in eastern Asia and eastern North America. Species identification for this genus is difficult if only using morphological evidence. L. bicolor, commonly called bush clover, is a deciduous shrub. Its native range is eastern Asia. It is hermaphrodite and is pollinated by insects. L. bicolor is an important ornamental plant due to its inflorescence. As a semi-woody perennial legume, it can fix nitrogen and may be used to prevent soil erosion. In this study, we reported the plastome of L. bicolor and resolved its phylogenetic relationship with other Lespedeza species.
Fresh leaves of L. bicolor were collected from Qianfo Mountain (Shandong, China; 36°37′N, 117°2′E). Voucher specimen (SD149) was deposited at College of Life Sciences, Shandong Normal University. Total genomic DNA was extracted by using the modified CTAB method, and was sequenced by the Novaseq platform at Novogene (Beijing, China). Plastome assembly was performed with Organelle Genome Assembler (OGA) as described in Qu, Fan, et al. (Citation2019). Plastome annotation was performed with Plastid Genome Annotator (PGA) (Qu, Moore, et al. Citation2019), with manual correction with Geneious v9.1.4. The annotated complete plastome was submitted to GenBank under accession number MT083894.
Complete plastome of L. bicolor was 148,930 bp in size with a canonical quadripartite structure, including two inverted repeats regions of 23,827 bp for each, a large single-copy region of 82,338 bp in length, and a small single-copy region of 18,938 bp in length. The overall guanine-cytosine (GC) content was 35.0%. A total of 112 unique genes was annotated in this plastome, including four rRNA genes, 30 tRNA genes, and 78 protein-coding genes. Among them, two protein-coding genes (clpP and ycf3) contained two introns, and eight protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, and rps16) and six tRNA genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained one intron.
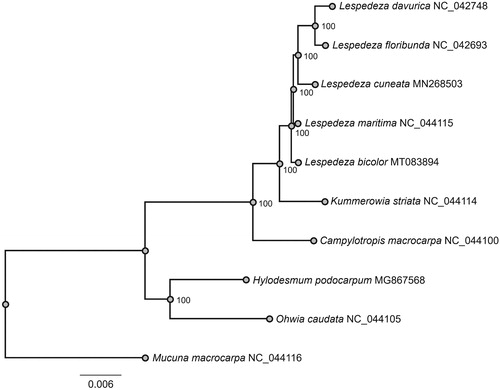
A maximum likelihood (ML) tree was reconstructed to determine the phylogenetic relationships of L. bicolor with other Lespedeza species by using RAxML v8.2.10 (Stamatakis Citation2014), including tree robustness assessment with 1,000 rapid bootstrap replicates, and GTRGAMMA substitution model. Alignment of 78 shared protein-coding genes was conducted by using MAFFT v7.313 (Katoh and Standley Citation2013). ML phylogenetic analysis showed that L. bicolor was sister to other Lespedeza species ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Qu XJ, Fan SJ, Wicke S, Yi TS. 2019. Plastome reduction in the only parasitic gymnosperm Parasitaxus is due to losses of photosynthesis but not housekeeping genes and apparently involves the secondary gain of a large inverted repeat. Genome Biol E. 11(10):2789–2796.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Xu B, Wu N, Gao XF, Zhang LB. 2012. Analysis of DNA sequences of six chloroplast and nuclear genes suggests incongruence, introgression, and incomplete lineage sorting in the evolution of Lespedeza (Fabaceae). Mol Phylogenet E. 62(1):346–358.