Abstract
In this study, we sequenced the first complete mitochondrial genome of Cryptalaus larvatus. The complete mitochondrial genome of C. larvatus was 15,876 bp with 29.80% GC containing 13 protein-coding genes, 22 transfer RNA (tRNA), two ribosomal RNA (rRNA), as well as and an AT-rich region. Phylogenetic analysis showed that the C. larvatus, Pyrophprus divergens, Ignelater luminosus, Hapsodrilus ignifer, and Dicronychus cinereus were clustered together. This study provides important information for the identification of this species and the study of genetic evolution with other species of Elateridae.
Elateridae, being called Click beetles, is a large family, including ∼400 genera and 9000 species, which is a kind of underground pest with widely distributed, serious harm, and strong concealment in the world (Lawrence Citation1982; Sagegami-Oba et al. Citation2007). In China, Cryptalaus larvatus is the dominant species of the Elateridae (Jiang Citation2003). Herein, the first complete mitochondrial genome of C. larvatus was sequenced and annotated, besides, its phylogenetic tree was constructed based on the neighbor-joining method. All voucher specimens were assigned with a unique code and deposited in the Key Laboratory of Integrated Pest Management in Ecological Forests, Fujian Province University, Fujian Agriculture and Forestry University (voucher no. CL-201907).
The samples of C. larvatus were caught by the traps of attractive substance from Minhou, Fujian Province, China (25°51′N, 119°22′E). The genomic DNA was extracted using TruSeq DNA Sample Preparation Kit according to the manufacturer’s instruction (Vanzyme, Nanjing, China) from the legs of samples. DNA quality and concentration were determined using Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA). Then, the DNA library with fragments of ∼300 bp in size was constructed by PCR amplification and the size selection by Agencourt AMPure XP-PCR Purification Beads (Beckman Coulter, CA, USA) and Agencourt SPRIselect Beads (Beckman Coulter, CA, USA). The multiple samples were mixed and sequenced by Illumina Hiseq 2500 (Genesky Biotechnologies Inc. Shanghai, China).
A total of 62,726,966 clean reads were obtained by filtration from the 63,763,234 raw reads. After the de novo assembly of metaSPAdes (Nurk et al. Citation2017), we obtained a 15,876 bp (GenBank accession No: MT118665) length complete mitochondrial genome of C. larvatus with 29.80% GC content. Three characteristics of protein-coding sequence, tRNA, and rRNA were obtained by the annotated result of mitoMaker (Bernt et al. Citation2013). And, tRNA genes were predicted by using tRNAscan (Lowe and Eddy Citation1997). A total of 37 genes were annotated, which contained 13 protein-coding genes, 22 transfer RNA (tRNA), and two ribosomal RNA (rRNA), there was also an AT-rich region.
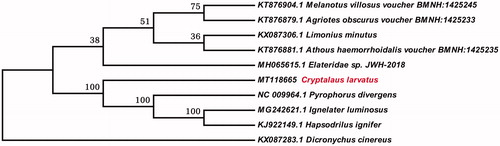
Phylogenetic tree was constructed using the neighbor-joining model with 1000 bootstrap replicates through FastTree software (Price et al. Citation2010) based on the genome sequence of C. larvatus and nine different species of Elateridae. The result showed that the C. larvatus, Pyrophprus divergens, Ignelater luminosus, Hapsodrilus ignifer and Dicronychus cinereus were clustered together, with high support value (). This study provides important information for the identification of this species and the study of genetic evolution with other species of Elateridae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in”NCBI”at https://www.ncbi.nlm.nih.gov/, reference number MT118665.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Jiang SH. 2003. Some questions of dominant species in the Chinese Elateridae Fauna. J Central South Forest Univ. 23:96–99.
- Lawrence JF. 1982. Coleoptera. In: Parker SP, editor. Synopsis and classification of living organisms. New York: McGraw-Hill; p. 482–553.
- Lowe TM, Eddy SR. 1997. tRNA scan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Price MN, Dehal PS, Arkin AP. 2010. FastTree2 – approximately maximum-likelihood trees for large alignments. PLOS One. 5(3):e9490.
- Sagegami-Oba R, Oba Y, Ohira H. 2007. Phylogenetic relationships of click beetles (Coleoptera: Elateridae) inferred from 28S ribosomal DNA: insights into the evolution of bioluminescence in Elateridae. Mol Phylogenet Evol. 42(2):410–421.