Abstract
Fragrant plantain lily (Hosta ventricosa) is an ornamental plant and widely cultivated in China due to its environmental adaptability. Here, the complete chloroplast genome of the H. ventricosa has been constructed from the Illumina sequencing data. The circular cp genome is 155,322 bp in size, and comprises a pair of inverted repeat (IR) regions of 24,912 bp each, a large single-copy (LSC) region of 84,113 bp, and a small single-copy (SSC) region of 20,782 bp. The total GC content is 38.0%, while the corresponding values of the LSC, SSC, and IR region are 36.2%, 32.5%, and 43.4%, respectively. The chloroplast genome contains 135 genes, including 88 protein-coding genes, 8 ribosomal RNA genes, and 39 transfer RNA genes. The neighbor-joining phylogenetic analysis showed a strong sister relationship with Hosta capitata in Hosta. Our findings provide a foundation for further investigation of chloroplast genome evolution in ornamental plant Hosta.
The genus Hosta in Asparagaceae, encompasses about 40 species and is widely distributed in temperate and subtropical regions and is cultivated in Europe as a garden plant (Liu et al. Citation2019). Hosta ventricosa is a perennial hermaphroditic plant that grows in a variety of habitats, including forest floors and edges, along the edges of roadways, and in open areas (Cao et al. Citation2011). The species is cultivated widely in China and western countries not only for its ornamental value, but also for its consideration as a medicinal herb for its pharmacological properties including anti-tumor, anti-inflammatory, anti-viral, antifungal, and anti-acetylcholinesterase activities (Li et al. Citation2012). In spite of its horticultural and medicinal values, there is are a few genetic and genomic studies available for breeding of this plant. Therefore, we reported the complete chloroplast genome (cp) of H. ventricosa based on Illumina sequencing data, which would be helpful for its evolution and genetics research.
Fresh leaves of H. ventricosa were collected from test field of Jilin Agriculture University (125°24′38″E, 43°48′39″N) for total genomic DNA extraction. The voucher specimen was preserved at the Herbarium of Jilin Agriculture University (accession number YZZE1909206). High-throughput DNA sequencing was conducted on the Illumina HiSeq 4000 Sequencing System (Illumina, CA, USA), and sequenced by Genesky Biotechnology (Shanghai, China). We assembled the cp genome using CLC Genomics Workbench v7.5 (CLC Bio, Aarhus, Denmark) as stated previously. A subset of 38.35 M trimmed reads were used for constructing the chloroplast genome by MITObim v1.8 (Hahn et al. Citation2013), with that of its congener Hosta minor (GenBank: NC_035999.1) as the initial reference genome. Finally, the validated complete chloroplast genome sequence was submitted to GenBank with the accession number MN901632.
The complete chloroplast genome of H. ventricosa is 155,322 bp in size with the mean coverage 327.9×, containing a pair of inverted repeat (IR) regions of 24,912 bp each, separated by a large single-copy (LSC) region of 84,113 bp and a small single-copy (SSC) region of 20,782 bp. The total GC content is 38.0%, while the corresponding values of the LSC, SSC, and IR regions are 36.2%, 32.5%, and 43.4%, respectively. This chloroplast genome harbors 135 functional genes, including 88 protein-coding genes, 39 tRNA genes, and 8 rRNA genes. Among them, 57 are involved in photosynthesis and 83 genes are involved in self replication. Six protein-coding genes, rpl2, rpl23, rps7, rps12, ndhB, and ycf2, were duplicate genes located in IR regions. Besides, 9 genes contain one intron, while chlp, rps12 and ycf3 harbor two introns. Summarily, the H. ventricosa cp genome is structurally similar to previously published ones (Lim et al. Citation2017; Jang et al. Citation2018).
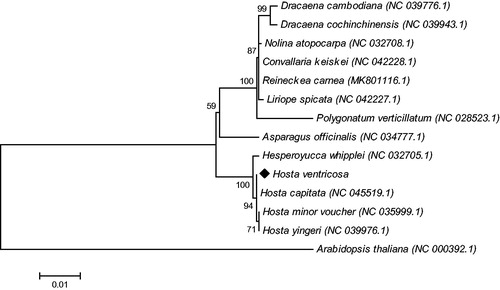
The phylogenetic tree was generated based on the complete cp genome of H. ventricosa and other 16 species (). The alignment was conducted using MAFFT (Katoh and Standley Citation2013). The phylogenetic tree was built using the neighbor-joining (NJ) method. The results showed that H. ventricosa was closely related to Hosta capitata in Hosta. Our findings provide a foundation for further investigation of chloroplast genome evolution in the ornamental plant Hosta.
Figure 1. The phylogenetic tree was constructed using chloroplast genome sequences of 13 species within the Asparagaceae family and Arabidopsis thaliana as an outgroup based on the neighbor-joining method using 500 bootstrap replicates. Chloroplast genome sequences used for this tree are Arabidopsis thaliana, NC_000932.1; Asparagus officinalis NC_034777.1; Convallaria keiskei NC_042228.1; Dracaena cambodiana NC_039776.1; Dracaena cochinchinensis NC_039943.1; Hesperoyucca whipplei, NC_032705.1; Hosta capitata, MH581151.1; Hosta minor voucher NC_035999.1; Hosta yingeri, NC_039976.1; Liriope spicata NC_042227.1; Reineckea carnea MK801116.1; Polygonatum verticillatum NC_028523.1.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Cao G, Xue L, Li Y, Pan K. 2011. The relative importance of architecture and resource competition in allocation to pollen and ovule number within inflorescences of Hosta ventricosa varies with the resource pools. Ann Bot. 107(8):1413–1419.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Jang Y, Park JY, Kang SJ, Park SH, Shim H, Lee TJ, Kang JH, Sung SH, Yang TJ. 2018. The complete chloroplast genome sequence of Hosta capitata (Koidz.) Nakai (Asparagaceae). Mitochondrial DNA Part B. 3(2):1052–1053.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li R, Wang MY, Li XB. 2012. Chemical constituents and biological activities of genus Hosta (Liliaceae). J Med Plants Res. 6(14):2704–2713.
- Lim CE, Kim S, Lee HO, Ryu S-A, Lee B-Y. 2017. The complete chloroplast genome of a Korean endemic ornamental plant Hosta yingeri SB Jones (Asparagaceae). Mitochondrial DNA Part B. 2017. 2(2):800–801.
- Liu ZX, Dong AR, Jia HQ, Zhou Y, Liu XF, Zhang XY. 2019. First report of pseudomonas syringae pv. syringae causing leaf blight on Hosta ventricosa in China. Plant Dis. 103(8):2123.
