Abstract
The complete chloroplast genome of Passiflora miniata Vanderpl. was reported in this study. The cp genome was 149,573 bp in length including two inverted repeats (IRs) of 25,012 bp, which were separated by LSC and SSC of 86,070 bp and 13,479 bp, respectively. The GC content was 37.1%. The genome encoded 106 functional genes, including 72 protein-coding genes, 30 tRNA genes, and four rRNA genes. The maximum likelihood phylogenetic tree indicated that P. miniata was recovered in the clade of subg. Passiflora and most related to P. vitifolia. This newly reported plastid genome will provide essential data for further study on the phylogeny and evolution of the genus and family.
Passiflora L. is the most species-rich genus in Passifloraceae with more than 600 species (Espinoza et al. Citation2018; Ma et al. Citation2019). This monophyletic group contains vines, lianas, shrubs and small trees that distributed mainly in Central and South America and few in Southeast Asia, Australia and the Pacific Islands (Killip Citation1938; De Wilde Citation1972). Passiflora is cultivated worldwide not only due to its horticultural value, but also for the edible fruits (Martin and Nakasone Citation1970). Despite Krosnick et al. (Citation2009, Citation2013) proposed five subgenera based on the phylogenetic relationship of Passiflora reconstructed by multiple loci of three genomes, the phylogenetic incongruence was detected by using the datasets of 64 plastid encoded protein genes (Rabah et al. Citation2019). Furthermore, Passiflora also occurred widespread genomic rearrangements (Rabah et al. Citation2019). In order to further explore the phylogenetic relationship within Passiflora as well as the pattern of its highly rearranged plastid genomes, we reported the complete chloroplast genome of P. miniata for the first time.
In this study, leaf materials were collected in the field of Mengla County, Yunnan, China (N 105°25′ E 21°41′) and immediately dried by silica gel for DNA extraction. Voucher specimen (HWH001) of this collection was deposited at IBK. Total genome DNA was extracted using the CTAB method (Doyle and Doyle Citation1987) and then sent to Majorbio Company (http://www.majorbio.com/, China) for next-generation sequencing using Illumina Hiseq 4000. We used Map to Reference function in Geneious R11 (Kearse et al. Citation2012) to exclude nuclear and mitochondrial reads using plastid genome of Passiflora quadrangularis (GenBank-MF807944) as reference. Putative chloroplast reads were used for de novo assembling construction using Geneious R11. Generated contigs were concatenated into larger ones using the Repeat Finder function in Geneious R11. The original data were again mapped to the larger contigs to extend their boundaries until all contigs were able to concatenate to one contig. The IR region was determined using the Repeat Finder function in Geneious R11 and was inverted and copied to obtain the complete chloroplast sequence. The annotation approach was performed followed Liu et al. (Citation2018) using same reference. The complete chloroplast genome of Passiflora miniata was 149,573 bp in length (GenBank-MT213977), the GC content was 37.1%. LSC and SSC contained 86,070 bp and 13,479 bp respectively, while IR was 25,012 bp in length. The plastid genome encoded 106 functional genes, including 72 protein-coding genes, 30 tRNA genes, and four rRNA genes.
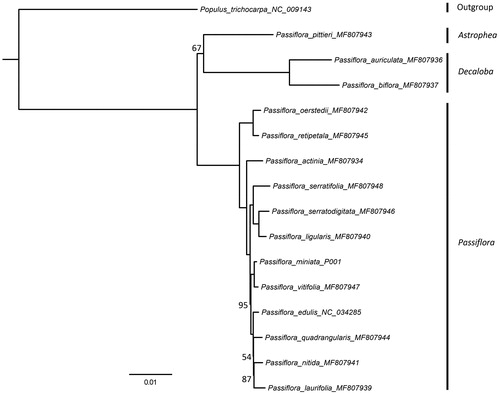
As the status of highly rearranged plastomes, it is unlikely to analyze the phylogenetic relationship by the dataset of complete genomes. Therefore, the maximum likelihood phylogenetic tree was reconstructed based on 64 plastid encoded protein genes including 15 species from Passiflora as ingroup and one species of Populus as outgroup (). The result was congruent with the study (Rabah et al. Citation2019) that subg. Astrophea as sister to subg. Decaloba and together are sister to subg. Passiflora. Although the result is still incongruence with the studies of Muschner et al. (Citation2012) and Krosnick et al. (2013) which can be explained as limited taxon sampling or plastid capture (Rabah et al. Citation2019), P. miniata falls into the clade of subg. Passiflora and is most closely related to P. vitifloia with strong support. It also exhibits the same plastome structure to those in subg. Pssiflora as concluded in Rabah et al. (Citation2019). This newly reported plastid genome will provide essential data for further study on the phylogeny and evolution of the genus Passiflora and of the family Passifloraceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- De Wilde W. 1972. The indigenous old world passifloras. Blumea. 20:227–250.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Espinoza TEB, Jørgensen PM, MacDougal JM. 2018. A taxonomic revision of Passiflora sect. Xerogona (Passifloraceae) using principal component analysis. Ann Missouri Bot Gard. 103(2):258–313.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Killip EP. 1938. The American species of Passifloraceae. Fieldiana Bot. 19:1–613.
- Krosnick SE, Ford AJ, Freudenstein JV. 2009. Taxonomic revision of Passiflora subgenus Tetrapathea including the monotypic genera Hollrungia and Tetrapathea (Passifloraceae), and a new species of Passiflora. Syst Bot. 34(2):375–385.
- Krosnick SE, Porter-Utley KE, MacDougal JM, Jørgensen PM, McDade LA. 2013. New insights into the evolution of Passiflora subgenus Decaloba (Passifloraceae): phylogenetic relationships and morphological synapomorphies. Syst Bot. 38(3):692–713.
- Liu H, He J, Ding C, Lyu R, Pei L, Cheng J, Xie L. 2018. Comparative analysis of complete chloroplast genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica revealing structural variations among genera in tribe Anemoneae (Ranunculaceae). Front Plant Sci. 9:1097.
- Ma XD, Yan LC, Krosnick SE, Zhu RB, Shi JP, Shen JY. 2019. Passiflora menghaiensis, a new species of Passifloraceae from Yunnan, China. Taiwania. 64(2):97–102.
- Martin FW, Nakasone HY. 1970. The edible species of Passiflora. Econ Bot. 24(3):333–343.
- Muschner VC, Zamberlan PM, Bonatto SL, Freitas LB. 2012. Phylogeny, biogeography and divergence times in Passiflora (Passifloraceae). Genet. Mol. Biol. 35(4 suppl 1):1036–1043.
- Rabah SO, Shrestha B, Hajrah NH, Sabir MJ, Alharby HF, Sabir MJ, Alhebshi AM, Sabir JSM, Gilbert LE, Ruhlman TA, et al. 2019. Passiflora plastome sequencing reveals widespread genomic rearrangements. J Syst Evol. 57(1):1–14.