Abstract
The complete mitochondrial genome of the Capsala pricei (Monogenea: Capsalidae) collected from the sailfish (Istiophorus platypterus) was sequenced by the next-generation sequencing method. The mitogenome is 13,851 bp in length, includes 12 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a control region, with the atp8 gene being absent. The overall content of A + T is 69.26%, which is significantly higher than the C + G content (30.74%). C shows the lowest frequency (13.42%) among four bases. Phylogeography analysis showed a close relationship between the genus Capsala and Neobenedenia with high bootstrap value supported.
Capsalids are ectoparasites of marine fish and some are important pathogens of fish in aquaculture and aquaria. Capsala pricei Hidalgo, 1959, a capsalid monogenean of the subfamily Capsalinae Johnston, 1929, is a frequently occurring pathogen of sailfish and swordfish. Feeding on mucus and epithelial cells, this parasite usually gives rise to inflammation, mucus hyperproduction and hemorrhage of their hosts (Paperna Citation1991). The morphological characters are the number and shape of marginal sclearites, the opening position of uterus, the shape of vitelline reservoir and the size of testes (Whittington et al. Citation2004). Though C. pricei has a high level of host-specification, the morphological characters can be easily affected by various factors such as the environmental temperature and salinity, the parasitizing position, the development degree of parasite (Whittington and Horton Citation1996). Thus, mitochondrial genome was sequenced in the present study to provide useful molecular markers for further species identification and phylogenetic research of C. pricei.
Samples of C. pricei were collected from the buccal cavity of the sailfish (Istiophorus platypterus) from northern South China Sea (115°19′51.6″E, 21°24′7.2″N). The specimen (CP20190401) of C. pricei was deposited in South China Sea Fisheries Research Institute, Chinese Academy of Fisheries Sciences. Total genomic DNA was extracted from the muscle tissue of C. pricei by using the Marine Animals Genomic DNA Extraction Kit (TIANamp, China). DNA extraction, genomic library construction, and next-generation sequencing were conducted following the previous publication reported by Loh et al. (Citation2015). Within the range of other monogenean mitogenomes, the complete mitogenome is 13,851 bp in length (GenBank accession no. MN746360), including 12 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a control region. In accord with the previously studied monogenean species (Plaisance et al. Citation2007; Huyse et al. Citation2008; Perkins et al. Citation2010; Kang et al. Citation2012; Zhang et al. Citation2014), the atp8 gene is absent and the mitochondrial genes of C. pricei are all transcribed from the heavy strand in the same direction. The A + T content for mitochondrial DNA of C. pricei is 69.26%, while the C + G content is 30.74%. Similar to all the reported monogenean species, C shows the lowest frequency (13.42%) among four bases.
Among the 12 protein-coding genes, one overlapping reading frame (32 nucleotides) was observed between nad4 and nad4L. The ATG start codon was used in 10 out of the 12 protein-coding genes (cox1, cox2, nad6, nad5, cox3, cytb, nad4, atp6, nad1 and nad3), while the start codon of nad4L and nad2 is GTG. It is important to note that 5 of 12 protein-coding genes are inferred to end with TAA terminated codon (cox2, nad5, cox3, nad1, and nad3), 6 end with theTAG terminated codon (cox1, nad6, cytb, nad4L, nad4, and atp6), and the gene nad2 ends with the codon T––. The longest gene is cox1 (1563 bp) in all protein-coding genes, whereas the shortest is nad4L gene (249 bp). The length of two ribosomal genes (rrnL and rrnS) is 527 bp and 737 bp, respectively. They are located between trnT and cox2, and are separated by trnC. Twenty-two tRNA genes (57–69 bp) were determined in the mitochondrial DNA of C. pricei, folding into cloverleaf secondary structures with normal base paring. One prominent none-coding region has been detected between the trnT and rrnL gene, and was determined to be 407 bp in length. Similar to those of some reported capsalid monogeneans (Perkins et al. Citation2010; Kang et al. Citation2012), the A + T content (75.18%) of the none-coding region was slightly higher than the overall A + T content (69.26%) of mt genome.
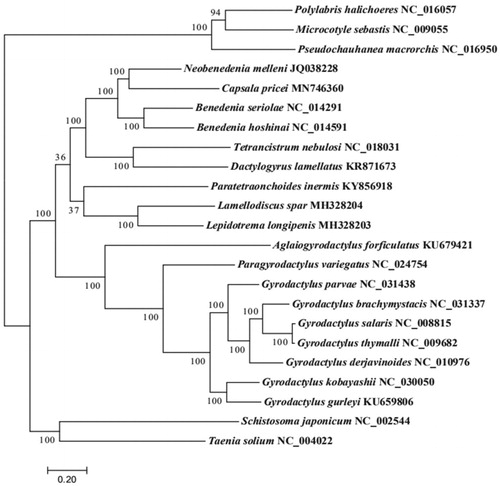
In order to validate the phylogenetic position of C. pricei, we constructed a maximum likelihood phylogenetic tree (1000 bootstrap replicates) using the complete mitochondrial sequences of 20 Monogenean species followed by the method of Liu et al. (Citation2020). Tenia solium and Schistosoma japonicum were used as outgroup to contrast the tree topology. Result shows that C. pricei can be unambiguously clustered with Neobenedenia melleni (), indicating a close relationship between the genus Capsala and Neobenedenia. The complete mitogenome of C. pricei provides useful molecular data for further phylogeography analyses of monogeneans.
Acknowledgements
We thank the editor and anonymous reviewers for their valuable comments on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov/nuccore/MN746360.1. We declared that materials described in the manuscript, including all relevant raw data, will be freely available to any scientist wishing to use them for noncommercial purposes, without breaching participant confidentiality.
References
- Huyse T, Buchmann K, Littlewood D. 2008. The mitochondrial genome of Gyrodactylus derjavinoides (Platyhelminthes: Monogenea) – a mitogenomic approach for Gyrodactylus species and strain identification. Gene. 417(1–2):27–34.
- Kang S, Kim J, Lee J, Kim S, Min G, Park J. 2012. The complete mitochondrial genome of an ectoparasitic monopisthocotylean fluke Benedenia hoshinai (Monogenea: Platyhelminthes). Mitochondrial DNA. 23(3):176–178.
- Liu Y, Shan BB, Yang CP, Zhao Y, Liu MT, Xie QJ, Sun DR. 2020. The complete mitochondrial genome of milk shark, Rhizoprionodon acutus (Ruppell 1837). Mitochondrial DNA Part B. 5(1):310–311.
- Loh KH, Shao KT, Chen HM, Chen CH, Chong VC, Loo PL, Shen KN, Hsiao CD. 2015. Next generation sequencing yields the complete mitochondrial genome of the Zebra moray, Gymnomuraena zebra (Anguilliformes: Muraenidae). Mitochondrial DNA. 1940–1744:1–2.
- Paperna I. 1991. Diseases caused by parasites in the aquaculture of warm water fish. Annu Revf Fish Dis. 1:155–194.
- Perkins EM, Donnellan SC, Bertozzi T, Whittington ID. 2010. Closing the mitochondrial circle on paraphyly of the Monogenea (Platyhelminthes) infers evolution in the diet of parasitic flatworms. Int J Parasitol. 40(11):1237–1245.
- Plaisance L, Huyse T, Littlewood DTJ, Bakke TA, Bachmann L. 2007. The complete mitochondrial DNA sequence of the monogenean Gyrodactylus thymalli (Platyhelminthes: Monogenea), a parasite of grayling (Thymallus thymallus). Mol Biochem Parasitol. 154(2):190–194.
- Whittington ID, Deveney MR, Morgan JAT, Chisholm LA, Adlard RD. 2004. A preliminary phylogenetic analysis of the Capsalidae (Platyhelminthes: monogenea: Monopisthocotylea) inferred from large subunit rDNA sequences. Parasitology. 128(5):511–519.
- Whittington ID, Horton MA. 1996. A revision of Neobenedenia Yamaguti, 1963 (Monogenea: Capsalidae) including a redescription of N. melleni (MacCallum, 1927) Yamaguti, 1963. J Nat Hist. 30(8):1113–1156.
- Zhang J, Wu XY, Li YW, Zhao MW, Xie MQ, Li AX. 2014. The complete mitochondrial genome of Neobenedenia melleni (Platyhelminthes: Monogenea): mitochondrial gene content, arrangement and composition compared with two Benedenia species. Mol Biol Rep. 41(10):6583–6589.