Abstract
We first determined the complete mitochondrial genome of Phymorhynchus sp. in this study. The total length of the mitochondrial genome of Phymorhynchus sp. was 15,631 bp, including 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and 1 no-control region. Phylogenetic analysis indicated that Phymorhynchus sp. was most closely related to the Typhlosyrinx sp., another species in the family Raphitomidae.
Phymorhynchus belongs to the family Raphitomidae, and is usually distributed in vent and seep environments (Zhang and Zhang, Citation2017). There were only two complete mitochondrial genomes of the species in Raphitomidae recorded in NCBI nr database. Here, we first sequenced and characterized the mitochondrial genome of Phymorhynchus sp., which was collected from vent filed on Central Indian Ridge, and examined the phylogenetic of Phymorhynchus sp. within Gastropoda.
The specie of Phymorhynchus sp. (GenBank accession number MT111940) was sampled from Kairei vent field on Central Indian Ridge (70.03992°E, −25.32041°N, 2438 m) on February 2019 and preserved in 95% ethanol. Total genomic DNA (voucher no. IDSSE-EEMB-L01) was extracted from foot muscle and immediately stored at –80 °C in the Institute of Deep-sea Science and Engineering, CAS. The complete mitochondrial genome of Phymorhynchus sp. was sequenced using the PCR technique. The gene annotation was done using the online program MITOS webserver (http://mitos.bioinf.unileipzig.de/index.py) (Bernt et al. Citation2013) and ORF Finder (http://www.bioinformatics.org/sms2/orf_find.html) (Rombel et al. Citation2002).
The size of mitogenome was 15,631bp, including 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and 1 no-control region, which overall nucleotide composition of the mitochondrial genome was 30.61% A, 38.57% T, 14.84% C, 15.99% G. All the mitogenome genes were encoded on the heavy strand (H-strand) except for seven tRNA genes.
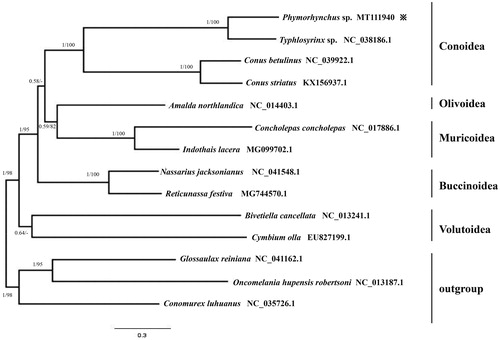
The phylogenetic analysis was based on 13 protein-coding genes of Phymorhynchus sp. and other 13 species. Oncomelania hupensis robertsoni, Glossaulax reiniana, Charonia lampas and Conomurex luhuanus were used as an outgroup. Phylogenetic reconstruction was performed using the Bayesian method (BI) and maximum likelihood method (ML) with the software MrBayes v.3.2 (Ronquist et al. Citation2012) and raxmlGUI v.1.3.1 (Silvestro and Michalak, Citation2012), respectively. Both BI and ML analyses produced identical tree topologies, and almost all the clades had a high support (BI posterior probabilities = 1 and bootstrap support = 100) (). Phylogenetic analysis revealed that Phymorhynchus sp. was in the clade of Conoidea, and compared with Volutoidea, Conoidea was most closely related to the Olivoidea, Muricoidea and Buccinoidea. This study will provide useful mitochondria information for further evolutionary analysis of this vent species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov/, GenBank accession number MT111940.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319
- Rombel IT, Sykes KF, Rayner S, Johnston SA. 2002. ORF-FINDER: a vector for high-throughput gene identification. Gene. 282(1-2):33–41.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systemat Biol. 61(3):539–542.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Zhang S, Zhang S. 2017. A new species of the genus Phymorhynchus (Neogastropoda: Raphitomidae) from a hydrothermal vent in the Manus Back-Arc Basin. Zootaxa. 4300(3):441–444.