Abstract
In order to improve our understanding of the characterization of Cheilinus fasciatus mitogenome and the classification status in Labridae, the full-length mtDNA of C. fasciatus was sequenced, which is 16,872 bp in length with the higher A + T content (53%). It is composed of 37 typical vertebrate mitochondrial genes and a noncoding control region (D-loop). Except for the ND6 gene and eight tRNAs, the rest of the genes are encoded on the heavy strand (H-strand). Subsequent phylogenetic analyses support that C. fasciatus belongs to the genus Cheilinus, which is closely related to two genera (Chlorurus and Scarus). The present study will provide important information for future understanding evolutionary theory, population molecular genetics and biological taxonomy of labrid fish.
The redbreast wrasse, Cheilinus fasciatus, belongs to the family Labridae that inhabits the reefs across the Indian and Pacific Ocean (Albutra et al. Citation2014). The family Labridae is the second-largest marine fish family, including more than 600 fish species belonging to 82 genera (Westneat and Alfaro Citation2005). Due to the structural and ecological diversity of this family, Labrid phylogenetic evolution has been a hot topic of research. Studies on the phylogenetic status in labrid species have been reported (Westneat and Alfaro Citation2005; Phillips et al. Citation2016). However, the phylogenetic analysis based on the Labridae mitogenome is unsystematic, hence only 19 Labridae mitogenomes have been published in GenBank. To better understand the phylogenetic status and genetic evolutions of Labridae mitogenome, the mitogenome of C. fasciatus was sequenced and its phylogenetic position among labrid species was analyzed.
The specimen used to sequence the whole mtDNA of the redbreast wrasse C. fasciatus originated from a wild individual, collected by ‘Nanfeng’ expedition ship of South China Sea Fisheries Research Institute, CAFS in the South China Sea of China (20°29.7′N, 117°56.3′E). Whole genomic DNA was extracted from the muscle tissue. The DNA library of C. fasciatus mtDNA is constructed and sequenced following Zhang et al. (Citation2015). The methods of genome annotation and genome sequence analysis were referred to Zhu et al. (Citation2017). All the specimens (accession number: Ssfri-F0092) and the genomic DNA were deposited in Tropical and Subtropical Marine Life Museum of South China Sea Fisheries Research Institute, CAFS, Guangzhou city, Guangdong Province, China.
The mitochondrial genome of C. fasciatus (GenBank accession no. KY115687.1) consists of 16,872 bp, and contains an essential set of typical vertebrate mitochondrial genes including 13 protein-coding genes, 22 tRNA genes, two rRNA genes and a noncoding control region (D-loop). Except for the ND6 gene and eight tRNAs, the rest of the genes are encoded on the heavy strand (H-strand). The arrangement of the 13 protein-coding genes in C. fasciatus is identical to that of the other sequenced labrid mitogenomes (Guo et al. Citation2019). The nucleotide composition of the C. fasciatus mitogenome is biased toward A + T (53.0%). Except for COXI (GTG), 12 PCGs initiate with a typical ATG start codon. Five genes (ND1, COXI, ATP8, ND4L, and ND5) are terminated in TAA codon and ND6 gene ended with TAG. The other genes are ended with incomplete stop codons (T or TA), expect ND4 with AGG mitochondrial specific stop codons also being found in some other fish (Prosdocimi et al. Citation2012).
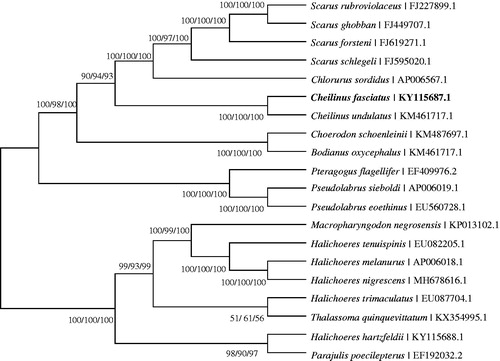
The phylogenetic trees using maximum parsimony (MP), maximum likelihood (ML), and neighbor-joining (NJ) based on the nucleotide of 13 PCGs from 20 labrid species () showed that C. fasciatus has a close relationship with C. undulatus and the genus Cheilinus is closely related to two genera (Chlorurus and Scarus), which is consistent with the previous studies (Zhu et al. Citation2017). The current work improves our understanding of the mitogenomic structure of C. fasciatus and evolutionary status of Labridae species.
The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov/nuccore/KY115687.1, reference number [KY115687.1].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
Additional information
Funding
References
- Albutra QB, Torres MAJ, Demayo CG. 2014. Qualitative and quantitative analysis of scale shape variation between different body regions of redbreast wrasse (Cheilinus fasciatus). Ann Biol Sci. 2(4):1–10.
- Guo HY, Zhang N, Zhu KC, Liu BS, Guo L, Yang JW, Zhang DC. 2019. Characterization of the complete mitochondrial genome sequence of orangeline wrasse Halichoeres hartzfeldii (Bleeker, 1852) with the phylogenetic relationships within the Labridae species. Mitochondrial DNA B. 4(2):3254–3255.
- Phillips GAC, Carleton KL, Marshall NJ. 2016. Multiple genetic mechanisms contribute to visual sensitivity variation in the Labridae. Mol Biol Evol. 33(1):201–215.
- Prosdocimi F, de Carvalho DC, de Almeida RN, Beheregaray LB. 2012. The complete mitochondrial genome of two recently derived species of the fish genus Nannoperca (Perciformes, Percichthyidae). Mol Biol Rep. 39(3):2767–2772.
- Westneat MW, Alfaro ME. 2005. Phylogenetic relationships and evolutionary history of the reef fish family Labridae. Mol Phylogenet Evol. 36(2):370–390.
- Zhang DC, Gong FH, Liu TT, Guo HY, Zhang N, Zhu KC, Jiang SG. 2015. Shotgun assembly of the mitochondrial genome from Fenneropenaeus penicillatus with phylogenetic consideration. Mar Genom. 24:379–386.
- Zhu KC, Wu N, Sun XX, Guo HY, Zhang N, Jiang SG, Zhang DC. 2017. Characterization of complete mitochondrial genome of fives tripe wrasse (Thalassoma quinquevittatum, Lay & Bennett, 1839) and phylogenetic analysis. Gene. 598:71–78.