Abstract
Dalbergia (Fabaceae) is a pantropical genus. Due to high economic and ecological values, many Dalbergia species were assessed as threatened taxa. In this study, we reported the complete plastome of two Dalbergia species, D. odorifera and D. oliveri, that was 155,838 bp and 156,074 bp in size, respectively. Comparative analyses showed Dalbergia plastomes were conserved in genome size, structure, and gene contents. Four nucleotide diversity hotspots of Dalbergia genomes were rpl32-ndhF, rpl32-trnLUAG, trnLUAA, and trnTACA-trnLUAA. Phylogenetic analysis revealed the sister relationship between D. odorifera and D. hainanensis, and between D. oliveri and D. assamica, respectively. The complete plastomes can provide the important information for investigations on conservation genetics and phylogenies of Dalbergia and Fabaceae.
Dalbergia L. f. (Faboideae, Fabaceae), comprises approximately 250 species as trees, shrubs, and woody climbers, which are widely distributed in tropical and sub-tropical regions around the world (Vatanparast et al. Citation2013; Hassold et al. Citation2016). Due to high economic and ecological values, all Dalbergia species have been included in appendices list of Wild Fauna and Flora by international trade regulations under the Convention on International Trade (CITES Citation2019). Meanwhile, 108 Dalbergia species are included in the IUCN Red List (IUCN Citation2020). Unfortunately, some Dalbergia species become endangered as the consequence of illegal timber trades. To enforce protective legislation and ensure effective conservation on Dalbergia species, the wood identity of the traded timber must be effective and accurate. Therefore, it would be highly desirable to develop reliable identification techniques being rapidly applied without specialists (Song et al. Citation2019). To date, DNA barcoding approach has been demonstrated as an effective molecular technique for species identification and community phylogenies (CBOL Plant Working Group Citation2009; Li et al. Citation2011; Kress Citation2017; Zeng et al. Citation2018). Chloroplast genomes of angiosperms have a circular structure and are composed of four regions, including two inverted repeat regions that are separated by a large single-copy (LSC) and a small single-copy (SSC) region (Palmer Citation1983; Wicke et al. Citation2011; Ruhlman and Jansen Citation2014). Chloroplast genomes of Dalbergia spp. range from 155,726 to 156,698 bp in size (Wariss et al. Citation2018; Deng et al. Citation2019; Song et al. Citation2019). In this study, we de novo assembled the complete chloroplast genome of two Dalbergia species, D. odorifera T.C.Chen and D. oliveri Gamble ex Prain, from China and Myanmar. The main goals of this study were to: (1) characterize the chloroplast genome of Dalbergia spp.; (2) assess high variable plastid regions for species identification and evolutionary investigations; and (3) reconstruct the plastid phylogenomics of Dalbergia, as well as Faboideae.
Fresh specimens of D. odorifera were collected from Xishuangbanna Tropical Botany Garden, Chinese Academy of Sciences, Yunnan, China (21°55′14″N and 101°16′34″E), and those of D. oliveri were collected from Ngaliak Reserve, Nay Pyi Taw Union Territory, Myanmar (19°92′68″N and 95°97′52″E). Voucher specimens of D. oliveri (P.P. Win et al. PPW-029) and D. odorifera (W-B Yu et al. Dai 166) were deposited at the Herbarium of Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences (HITBC).
Genomic DNAs were extracted from silica gel dried leaves using a modified CTAB method (Doyle and Doyle Citation1987). The purified DNA was fragmented to approximately 350 bp in size for library construction, and 150 bp pair-end reads were generated by Illumina NovaSeq 6000 (Annoroad, Beijing, China). The raw reads were de novo assembled using GetOrganelle toolkit (Jin et al. Citation2018). The plastomes were annotated using CPGAVAS2 (Shi et al. Citation2019), then manually adjusted in Geneious (Kearse et al. Citation2012). Fifty-two plastomes of Faboideae were achieved from GenBank, and Cercis canadensis L. was chosen as outgroup. The whole plastome sequences with on IR region were aligned using MAFFT (Katoh and Standley Citation2013), gaps were trimmed by trimAl (Capella-Gutiérrez et al. Citation2009) using the command ‘-gt 0.5 cons 50′. The maximum likelihood tree was reconstructed by RAxML (Stamatakis Citation2014) using GTRGAMMAI model with 1000 bootstrap replicates on CIPRES (https://www.phylo.org/). The tree was visualized with the TreeGraph (Stover and Muller Citation2010). The dynamic nucleotide diversity (π) of seven Dalbergia plastomes was estimated using DnaSP (Rozas et al. Citation2017) by a window size 500 bp and step size 250 bp. The region was considered as the divergence hotspot, when values of π were higher than 0.04%.
Two plastomes of Dalbergia were a typical quadripartite structure and 156,074 bp (D. odorifera, MT009405) and 155,838bp (D. oliveri, MT009406) in length. The LSC region for D. odorifera and D. oliveri was 85,766 bp and 85,829 bp, respectively, the SSC region was 18,841 bp and 18,686 bp, respectively, and the IR regions were 25,702 and 25,693 bp, respectively. GC contents were 36.1%. The genomes had 111 unique genes, including 77 protein coding genes (missing infA, rpl22, and ycf15), 30 transfer RNA (tRNA) genes, and four ribosomal RNA (rRNA). Four nucleotide diversity hotspots, rpl32-ndhF, rpl32-trnLUAG, trnLUAA, and trnTACA-trnLUAA, were identified across the plastomes of Dalbergia, which are informative regions as DNA barcodes for molecular authentication of Dalbergia species.
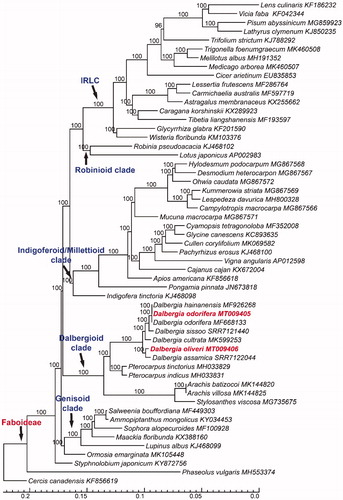
Phylogenetic analysis showed that the backbone of Faboideae was well resolved, and five major clades were fully supported (). Dalbergioid clade included four genera, showing a sister relationship between Stylosanthes+Arachis and Dalbergia+Pterocarpus. Dalbergia species formed a clade with 100% bootstrap values, and D. assamica Pittier and D. oliveri formed a clade, which was sister to the remaining Dalbergia species (). D. sissoo was sister to D. oliveri+D. hainanensis Merr. & Chun. Therefore, this study demonstrated that the whole chloroplast genome sequences can reconstruct a robust phylogeny of Dalbergia, as well as Faboideae.
Acknowledgments
We are grateful to the physical supports from HPC Platform of the Public Technology Service Center and Department of Gardening and Horticulture, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The sequencing data using in this manuscript are deposited in the GenBank sequence database. The data were collected without violation of the protection of human subjects, or other valid ethical, privacy, or security concerns.
Additional information
Funding
References
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- CBOL Plant Working Group. 2009. A DNA barcode for land plants. Proc Natl Acad Sci USA. 106:12794–12797.
- CITES. 2019. The checklist of CITES species, the convention on international trade in Endangered species of wild Fauna and Flora. http://checklist.cites.org/#/en.
- Deng C-Y, Xin G-L, Zhang J-Q, Zhao D-M. 2019. Characterization of the complete chloroplast genome of Dalbergia hainanensis (Leguminosae), a vulnerably endangered legume endemic to China. Conservation Genet Resour. 11(1):105–108. doi:10.1007/s12686-017-0967-y.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Hassold S, Chen CA, Lowry PP, Bauert MR, Razafintsalama A, Ramamonjisoa L, Widmer A. 2016. DNA barcoding of Malagasy rosewoods: towards a molecular identification of CITES-listed Dalbergia species. PLoS One. 11(6):e0157881.
- IUCN. 2020. The IUCN red list of threatened species, version 2020-1. www.iucnredlist.org.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv. 9:138.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kress WJ. 2017. Plant DNA barcodes: applications today and in the future. J Syst Evol. 55(4):291–307.
- Li D-Z, Gao L-M, Li H-T, Wang H, Ge X-J, Liu J-Q, Chen Z-D, Zhou S-L, Chen S-L, Yang J-B, et al. 2011. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci USA. 108(49):19641–19646.
- Palmer JD. 1983. Chloroplast DNA exists in two orientations. Nature. 301(5895):92–93.
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 34(12):3299–3302.
- Ruhlman TA, Jansen RK. 2014. The plastid genomes of flowering plants. In: Maliga P, editor. Chloroplast biotechnology: methods and protocols. Totowa (NJ): Humana Press; p. 3–38.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Song Y, Zhang Y, Xu J, Li W, Li M. 2019. Characterization of the complete chloroplast genome sequence of Dalbergia species and its phylogenetic implications. Sci Rep. 9(1):20401.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Stover BC, Muller KF. 2010. Tree Graph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 11:7.
- Vatanparast M, Klitgård BB, Adema FACB, Pennington RT, Yahara T, Kajita T. 2013. First molecular phylogeny of the pantropical genus Dalbergia: implications for infrageneric circumscription and biogeography. S Afr J Bot. 89:143–149.
- Wariss HM, Yi T-S, Wang H, Zhang R. 2018. Characterization of the complete chloroplast genome of Dalbergia odorifera (Leguminosae), a rare and critically endangered legume endemic to China. Conservation Genet Resour. 10(3):527–530. doi:10.1007/s12686-017-0866-2.
- Wicke S, Schneeweiss G, dePamphilis C, Müller K, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76(3–5):273–297.
- Zeng C-X, Hollingsworth PM, Yang J, He Z-S, Zhang Z-R, Li D-Z, Yang J-B. 2018. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods. 14(1):43.