Abstract
Andrographis paniculata is widely distributed and used in tropical and subtropical regions of Asia. To enhance the development of its medicinal value, we reported and characterized the complete chloroplast genome sequence of A. paniculata in an effort to provide genomic resources useful to promoting its medicinal value and systematic research. The chloroplast genome of Andrographis paniculata was found to possess a total length 150,249 bp with the typical quadripartite structure of angiosperms, respectively. It contained a large single-copy (LSC) region of 82,460 bp, a small single-copy (SSC) region of 17,111 bp and separated by a pair of inverted repeat (IR) regions of 25,339 bp. The chloroplast genome of Andrographis paniculata is composed of 132 individual genes, consisting of 87 protein-coding genes, 37 transfer RNA genes and 8 ribosomal RNA genes. The overall G-C content of the chloroplast genome is 38.4%, A of 45,852 bp (30.5%), T of 46,873 bp (31.2%), C of 29,281 bp (19.5%) and G of 18.8% (28,243 bp). Phylogenetic tree analysis result shown that Andrographis paniculata was clustered to Strobilanthes cusia by the maximum-likelihood (ML) method in this study.
Andrographis paniculata is an herbaceous plant belonging to the family Acanthaceae and subtropical regions of Asia countries, for example China, India, Thailand and Malaysia (Lim et al. Citation2012). Andrographis paniculata known as ‘Chuan-Xin-Lian’ in Chinese, which is well known and historically documented in both traditional Chinese medicine (TCM) in China (Pholphana et al. Citation2013). Andrographis paniculata can be used to treat a variety of diseases as the traditional Chinese medicine in China, such as diarrhea, fevers, laryngitis, gastric infections and upper respiratory tract infections (Sun et al. Citation2019). So, to enhance the development of its medicinal value, we reported and characterized the complete chloroplast genome sequence of A. paniculata in an effort to provide genomic resources useful to promoting its medicinal value and systematic research. This study also can provide a useful resource for the conservation genetics of this species and the phylogenetic studies of traditional Chinese medicine plant.
The plant material of Andrographis paniculata was collected from the herb market near Zhejiang Chinese Medical University (Hangzhou, Zhejiang, China in 119.89E, 30.09 N). We used the modified CTAB method to extract the chloroplast genome DNA, which was deposited at Zhejiang Chinese Medical University (No. ZJCMU-006). After, we used to remove the low-quality reads by FastQC (Andrews Citation2015), the sequence of A. paniculata was analyzed and assembled by MitoZ (Meng et al. Citation2019). The annotation was corrected with DOGMA (Wyman et al. Citation2004). The chloroplast genome sequence had been submitted to NCBI GenBank database under accession number of MN0458992, respectively.
The chloroplast genome of Andrographis paniculata was found to possess a total length 150,249bp with the typical quadripartite structure of angiosperms, respectively. The chloroplast genome composed of 82,460 bp of LSC region, 17,111 bp of SSC region and 25,339 bp of a pair of IR regions.
The chloroplast genome of Andrographis paniculata is composed of 132 individual genes, consisting of 87 protein-coding genes, 37 transfer RNA genes and 8 ribosomal RNA genes. Among the total 18 genes on the region, 7 protein-coding genes, 7 transfer RNA genes and 4 ribosomal RNA genes were duplicated on the IR regions. The overall G-C content of the chloroplast genome is 38.4%, A of 45,852 bp (30.5%), T of 46,873 bp (31.2%), C of 29,281 bp (19.5%) and G of 18.8% (28,243 bp).
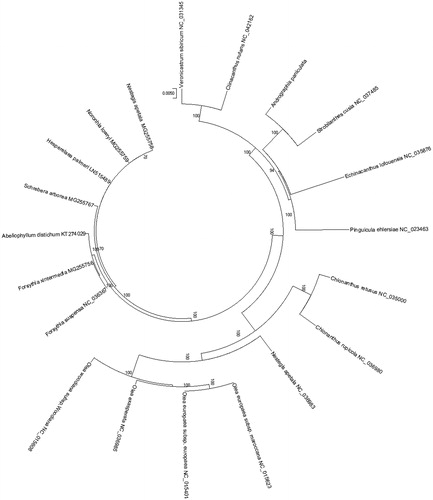
We used MEGA X (Kumar et al. Citation2018) with 2000 bootstraps under the best model to reconstruct the maximum-likelihood (ML) phylogenetic tree. The phylogenetic tree based on the chloroplast genome of 20 plant species sequences to study the evolutionary relationship with Andrographis paniculata. At the last, we used iTOL (Letunic and Bork Citation2016) to edit the ML phylogenetic tree. The phylogenetic tree analysis result indicates that Andrographis paniculata is closer to Strobilanthes cusia in this study (), the most nodes in the phylogenetic ML tree were strongly supported. The complete genome sequence of Andrographis paniculata can provide a useful resource for the conservation genetics of this species and enhance the development of its medicinal value in China.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v4: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245.
- Lim JCW, Chan TK, Ng DS, Sagineedu SR, Stanslas J, Wong WF. 2012. Andrographolide and its analogues: versatile bioactive molecules for combating inflammation and cancer. Clin Exp Pharmacol Physiol. 39(3):300–310.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Pholphana N, Rangkadilok N, Saehun J, Ritruechai S, Satayavivad J. 2013. Changes in the contents of four active diterpenoids at different growth stages in Andrographis paniculata (Burm.f.) Nees (Chuanxinlian). Chin Med.
- Sun W, Leng L, Yin QG, Xu MM, Huang MK, Xu ZC, Zhang YJ, Yao H, Wang CX, Xiong C, et al. 2019. The genome of the medicinal plant Andrographis paniculata provides insight into the biosynthesis of the bioactive diterpenoid neoandrographolide. Plant J. 97(5):841–857.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.