Abstract
The complete chloroplast genome of Youngia gracilipes was identified. Here, we sequenced and annotated complete chloroplast genome of Youngia gracilipes. The whole genome length is 152,811 bp, including a large single-copy (LSC) region with length of 84,337 bp, a small single-copy (SSC) region with length of 18,392 bp, a pair of inverted repeat (IR) regions with length of 25,041 bp. A total of 114 unique genes was found, of which 80 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Simultaneously, molecular phylogenetic analysis suggested that Youngia gracilipes was the closest to Youngia japonica.
The genus Youngia Cass. have ca. 30 species, mainly distributed in East Asia (Shih and Kilian Citation2011; Babcock and Stebbins Citation1937). It highly diversified in China, and many new species were discovered recently, such as Youngia baoxingensis, Youngia jiulongensis, and Youngia zhengyiana (Chen Citation2018; Peng et al. Citation2017; Deng et al. Citation2014). Youngia gracilipes is characterized by the longest outer phyllary being about one-third of the inner and no crested or corniculate appears at inner phyllaries (Shih Citation1997; Shih and Kilian Citation2011). This species was classified into a new genus Pseudoyoungia by Maity and Maiti (Citation2010). However, subsequent pollen morphological and molecular studies did not support the isolation of this new genus (Shih and Kilian Citation2011; Peng et al. Citation2013; Peng et al. Citation2014; Deng et al. Citation2014). In addition, Y. gracilipes grows in alpine meadows at an altitude of 2700–4800 m (Shih and Kilian Citation2011), which makes specimen collection difficult. This species was found in a recent field scientific expedition. Plus Y. gracilipes was not used in recent Asteraceae phylogenetic study of plastome genome (Lin et al. Citation2019). Therefore, we reported the complete plastome genome of Y. gracilipes and reconstructed phylogenetic tree with related species for the convenience of a detailed study of the phylogeny.
The materials of molecular experiments of Y. gracilipes were collected from Têwo Zong, Gansu Province, China (N34°15′17.99′′ and E103°06′52.35′′). The voucher specimens were deposited in KUN (Herbarium, Kunming Institute of Botany, Kunming, China, CAS; ZJW4887). The Y. gracilipes was sequenced based on next-generation sequencing technology (NGS) at the Beijing Novogene Bioinformatics Technology Co., Ltd, Beijing, China. The filtered clean reads were assembled using NOVOPlasty version 3.3 (Dierckxsens et al. Citation2017) with sequence of Ribulose-1,5-bisphosphate (RuBP) as a seed for starting. The complete chloroplast genome was annotated using Plastid Genome Annotator (PGA) (Qu et al. Citation2019). The annotated Amborella trichopoda and Soroseris umbrella plastome sequences were used as references, and all annotated results were manually corrected in Geneious version 9.0.2 (Kearse et al. Citation2012). At last, the assembled sequence was submitted to NCBI with annotations (NCBI accession number: MT267485).
The complete chloroplast genome of Y. gracilipes is 152,811 bp in size, which is divided into four parts by two inverted repeat (IR) sequences, containing an LSC region (84,337 bp), and SSC region (18,392 bp), and a pair of IR regions (25,041 bp). The GC content of whole genome is 37.8%. In addition, the values of LSC region, SSC region, and IR region are 35.9, 31.5, and 43.3%, respectively. A total of 114 unique genes were identified, including 80 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Among these, 12 protein-coding genes (ndhA, rps12, ndhB, rpl2, rpl16, petD, petB, clpP, ycf3, atpF, rpoC1, and rps16) and 6 tRNA genes (trnA-UGC, trnI-GAU, trnV-UAC, trnL-UAA, trnG-UCC, and trnK-UUU) are intron-containing genes.
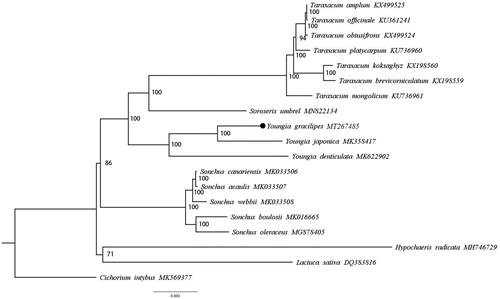
The phylogenetic status was confirmed by maximum likelihood (ML) method with 77 shared CDS sequences. All sequences including 19 species (Lv et al. Citation2020) from the Cichorieae were aligned using MAFFT version 7.308 (Katoh et al. Citation2002). The GTR + G + I was selected as best-fit model of DNA evolution by jModelTest version 2.1.10 (Darriba et al. Citation2012). Phylogenetic tree was reconstructed using RAxML version 8.2.10 (Alexandros Citation2014) with 1000 bootstrap replicates. Figtree version 1.4.4 (Rambaut Citation2018) was used to view the phylogenetic tree. Ultimately, the phylogenetic analysis results indicated that Y. gracilipes was the closest to Youngia japonica with 100% bootstrap values ().
Data availability
The data that support the findings of this study are available in [GenBank] at [https://www.ncbi.nlm.nih.gov/genbank/], reference number [MT267485].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Alexandros S. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Babcock EB, Stebbins GL. 1937. The genus Youngia. Washington (DC): Carnegie Institution of Washington; p. 1–106.
- Chen YS. 2018. Youngia baoxingensis, a new species of Asteraceae (tribe Cichorieae) from Sichuan, China. Phytotaxa. 382(2):239–242.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772.
- Deng T, Zhang JW, Zhu XX, Zhang DG, Nie ZL, Sun H. 2014. Youngia zhengyiana (Asteraceae, Crepidinae), a new species from south China, with notes on the systematics of Youngia inferred from morphology and nrITS phylogeny. Phytotaxa. 170(4):259–268.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lin N, Zhang X, Deng T, Zhang JW, Meng AP, Wang HC, Sun H, Sun YX. 2019. Plastome sequencing of Myripnois dioica and comparison within. Asteraceae Plant Divers. 41(5):315–322.
- Lv ZY, Zhang JW, Chen JT, Li ZM, Sun H. 2020. The complete chloroplast genome of Soroseris umbrella (Asteraceae). Mitochondrial DNA Part B. 5(1):637–638.
- Maity D, Maiti GG. 2010. Taxonomic delimitation of the genus Tibetoseris Sennikov and the new genus Pseudoyoungia of the Compositae-Cichorieae from Eastern Himalaya. Compositae Newsletter. 48:22–42.
- Peng YL, Gao XF, Peng L. 2013. Pollen morphology of Youngia and six related genera (Asteraceae: Cichorieae) and its systematic significance. Phytotaxa. 139(1):39–62.
- Peng YL, Gao XF, Zhang LB. 2017. Youngia jiulongensis (Crepidinae, Cichorieae, Asteraceae), a New Species from Sichuan, China. Novon A J Bot Nomenclature. 25(3):298–301.
- Peng YL, Zhang Y, Gao XF, Tong LJ, Li L, Li RY, Zhu ZM, Xian JR. 2014. A phylogenetic analysis and new delimitation of Crepidiastrum (Asteraceae, tribe Cichorieae). Phytotaxa. 159(4):241–255.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Rambaut A. 2018. FigTree version 1.4.4. Computer program and documentation distributed by the author. http://tree.bio.ed.ac.uk/software/figtree/
- Shih C. 1997. Compositae. In: Ling Y, Shih C, editors. Flora reipublicae popularis sinicae. Vol. 80. Beijing, China: Science Press; p. 106–266.
- Shih C, Kilian N. 2011. Youngia. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 20–21. Beijing, China: Science Press; St. Louis (MO): Missouri Botanical Garden Press; p. 252–264.