Abstract
In this study, we sequenced the full mitochondrial genome of the yellow prawn-goby, Cryptocentrus cinctus, an abundantly available fish in in the Western Pacific, which may be a useful resource for understanding the molecular evolutionary history of gobiine-like gobiids. The mitochondrial genome of C. cinctus is 16,514 bp long, and contains 13 protein-coding genes (PCGs) along with 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and a D-loop region, arranged similarly to those of Gobiidae species. A phylogenetic analysis of Gobiidae species indicates close relationships to the gobiine-like genus Oxyurichthys and Yongeichthys. The complete mitochondrial genome of C. cinctus will allow further understanding for analyzing evolutionary process in Gobiidae.
The teleost family Gobiidae is one of the largest vertebrate families that occupies a wide range of geographical distribution with tropical and temperate habitats, ecological flexibility, variety in morphology, and special adaptation (Patzner Citation2011; Tornabene et al. Citation2013). Since Gobiidae has complex evolutionary history, extensive genetic markers has been phylogenetically employed with evidences of geographical and morphological characteristics (Thacker Citation2009; Thacker and Roje Citation2011; Agorreta et al. Citation2013; Adrian-Kalchhauser et al. Citation2017). The yellow prawn-goby, Cryptocentrus cinctus (Herre 1936) also known as yellow watchman goby, is native to sandy areas of the Western Pacific. It has a fascinating habitat that shares a burrow with alpheid shrimps as mutualistic symbiosis. However, research on the phylogenetic relationship and evolutionary history of the genus Cryptocentrus remains to be still explored due to insufficient genomic information.
An individual fish was collected from East China Sea (28°19′31.1″N 126°03′33.7″E) in June 2012, and DNA was extracted from muscle tissue using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany), following the manufacturer’s protocol. The specimen is stored in the Research Institute of Basic Sciences of Incheon National University (Incheon, South Korea) with a specimen ID as 2013-gobiidae007 (Nam and Rhee Citation2020). A genomic sequencing library was prepared with total genomic DNA (1 μg) using the TruSeq DNA Sample Preparation Kit according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). Sequencing was completed with Illumina HiSeq-X platform (Illumina) by Phyzen (Seoul, South Korea). Assembly was conducted in CLC Assembly Cell package (version 4.2.1) using the CLC de novo assemble algorithm. Finally, a consensus mitogenome information was produced from a circular alignment of contiguous sequences. Additional PCR procedures were conducted to confirm the nucleotide sequence of the putative D-loop region. Overall sequences were reannotated using the MITOS web-based software (Bernt et al. Citation2013) and detailed annotation were further conducted with NCBI-BLAST (http://blast.ncbi.nlm.nih.gov).
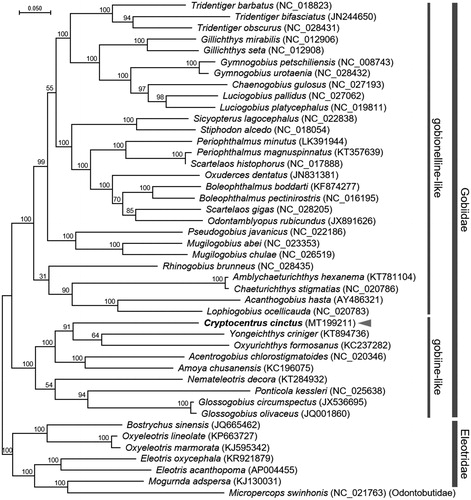
The mitochondrial genome of C. cinctus is 16,514 bp long (Accession no. MT199211). It contains the typical set of 13 PCGs, 22 tRNAs, 2 rRNAs, and a D-loop region. The genomic content and structure are very similar to other Gobiidae mitogenomes. A phylogenetic analysis was conducted by employing a maximum-likelihood (ML) method in the PhyML 2.4.5 (Guindon and Gascuel Citation2003) with 1000 bootstrap replicates under a HKY + G + I substitution model. Phylogenetic relationships based on the concatenated set of 13 PCGs’ sequences of the 36 Gobiidae species with 6 species from the family Eleotridae and a species from the family Odontobutidae indicates that the C. cinctus mitogenome is most closely related to the genus Oxyurichthys and Yongeichthys within a gobiine-like clade (). In conclusion, the complete C. cinctus mitogenome will be a useful resource for comparative analysis of the phylogenetic distance and geographical distribution in gobiine-like gobiids.
Figure 1. Maximum-likelihood (ML) phylogeny of 28 species of gobionelline-like and 8 species of gobiine-like gobiids based on the 13 concatenated nucleotide sequences of the entire protein-coding genes (PCGs). Six species from the family Eleotridae and a species from the family Odontobutidae were incorporated to build a reliable phylogeny tree. Node labels indicate posterior probability. DDBJ/EMBL/Genbank accession numbers for published sequences are incorporated.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, accession number MT199211.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Adrian-Kalchhauser I, Svensson O, Kutschera VE, Alm Rosenblad M, Pippel M, Winkler S, Schloissnig S, Blomberg A, Burkhardt-Holm P. 2017. The mitochondrial genome sequences of the round goby and the sand goby reveal patterns of recent evolution in gobiid fish. BMC Genomics. 18(1):177.
- Agorreta A, San Mauro D, Schliewen U, Van Tassell JL, Kovačić M, Zardoya R, Rüber L. 2013. Molecular phylogenetics of Gobioidei and phylogenetic placement of European gobies. Mol Phylogenet Evol. 69(3):619–633.
- Bernt A, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biol. 52(5):696–704.
- Nam S-E, Rhee J-S. 2020. Complete mitochondrial genome of the fire goby, Nemateleotris magnifica (Perciformes, Gobiidae). Mitochondrial DNA. 5(2):1894–1896.
- Patzner RA. 2011. The biology of gobies. 1st ed. Boca Raton: Science Publishers;
- Thacker CE. 2009. Phylogeny of Gobioidei and placement within acanthomorpha, with a new classification and investigation of diversification and character evolution. Copeia. 2009(1):93–104.
- Thacker CE, Roje DM. 2011. Phylogeny of Gobiidae and identification of gobiid lineages. Syst Biodiv. 9(4):329–347.
- Tornabene L, Chen Y, Pezold F. 2013. Gobies are deeply divided: phylogenetic evidence from nuclear DNA (Teleostei: Gobioidei: Gobiidae). Syst Biodivers. 11(3):345–361.
