Abstract
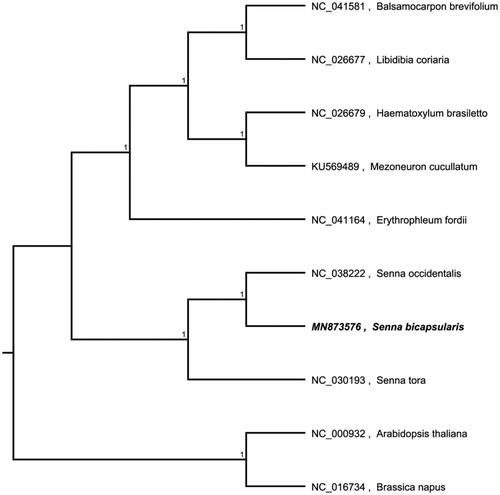
The complete chloroplast genome sequence of Senna bicapsularis was characterized from Illumina pair-end sequencing. The chloroplast genome of S. bicapsularis was 161,056 bp in length, containing a large single-copy region (LSC) of 90,416 bp, a small single-copy region (SSC) of 18,538 bp, and two inverted repeat (IR) regions of 26,051 bp. The overall GC content is 36.20%, while the correponding values of the LSC, SSC, and IR regions are 64.5%, 69.4%, and 60.2%, respectively. The genome contains 129 complete genes, including8 rRNAs, 37 tRNAs and 84 protein coding genes. A phylogenetic analysis showed that Senna tora and Erythrophleumfordiiform the basis of the produced evolutionary tree. S.bicapsularis and S. occidentalis, which belongto the group Cassia, share the closest relationship. The analysis of the cp genome of S.bicapsularisprovides crucial genetic information for further studies of this precious species and the taxonomy, phylogenetics and evolution of Cassia.
Senna bicapsularis (family: Fabaceae) is an ornamental plant belonging to Cassia species which is widely distributed in South American and tropical countries. Traditionally, plants belonging to Cassia species are believed to possess medicinal values. The wood of S. bicapsularis can be used to make paper pulp and its fruits are said to be edible (Mak et al. Citation2013). Nevertheless, its genetic background and resources have not been widely studied. Polymorphic chloroplast microsatellite markers designed based on a cp genome analysis can be utilized to comprehend the levels and patterns of the geographical structure and genetic diversity of S. bicapsularis, and this information can subsequently be used to formulate an effective protection strategy.
A single individual of S. bicapsularis was used as a sampling object from the China West Normal University (106°08′E; 30°78′N) in Nanchong. Fresh leaves of the individuals were collected and flash-frozen in liquid nitrogen and then stored in a refrigerator (–80 °C) until DNA extraction. The voucher specimen (SJJM001) was laid in the Herbarium of China West Normal University and the extracted DNA was stored in the –80 °C refrigerator of the Key Laboratory of Southwest China Wildlife Resources Conservation. We extracted total genomic DNA from 25 mg silica-gel-dried leaf using a modified CTAB method (Doyle Citation1987). The Illumina HiSeq 2000 platform (Illumina, San Diego, CA) was used to perform the genome sequence. We used the software MITObim 1.8 (Hahn et al. Citation2013) and metaSPAdes (Nurk et al. Citation2017) to assemble chloroplast genomes. We used S. occidentalis (GenBank: NC_038222) as a reference genome. We annotated the chloroplast genome with the software DOGMA (Wyman et al. Citation2004), and then corrected the results using Geneious 8.0.2 (Campos et al. Citation2016) and Sequin 15.50 (http://www.ncbi.nlm.nih.gov/Sequin/).
The complete chloroplast genome sequence of S. bicapsularis (GenBank number: MN873576) was characterized from Illumina pair-end sequencing. The chloroplast genome of S. bicapsularis was 161,056 bp in length, containing a large single-copy region (LSC) of 90,416 bp, a small single-copy region (SSC) of 18,538 bp, and two inverted repeat (IR) regions of 26,051 bp. The overall GC content is 36.20%, while the corresponding values of the LSC, SSC, and IR regions are 64.5%, 69.4%, and 60.2%, respectively. The genome contains 129 complete genes, including8 rRNAs, 37 tRNAs and 84 protein-coding genes. A phylogenetic analysis showed that S.bicapsularis and S. occidentalis, which belong to the group Cassia, share the closed relationship. The analysis of the cp genome of S.bicapsularis provides crucial genetic information for further studies of this precious species and the taxonomy, phylogenetics and evolution of Cassia.
We used the complete chloroplast genomes sequence of S. bicapsularis and 9 other related species and Brassica napus and Arabidopsis thaliana as an outgroup to construct the phylogenetic tree. The 10 chloroplast genome sequences were aligned with MAFFT (Katoh and Standley Citation2013), and then the neighbour-joining tree was constructed by MEGA 7.0 (Kumar et al. Citation2016). The results confirmed that S.bicapsularis was clustered with S. occidentalis ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The GenBank accession number for the cp genome sequence of S. bicapsularis is MN873576 and the DOI is https://www.ncbi.nlm.nih.gov/nuccore/MN873576.
Additional information
Funding
References
- Campos FS, Kluge M, Franco AC, Giongo A, Valdez FP, Saddi TM, Brito WMED, Roehe PM. 2016. Complete genome sequence of porcine parvovirus 2 recovered from swine sera. Genome Announc. 4(1):e01627–01615.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Mak YW, Chuah LO, Ahmad R, Bhat R. 2013. Antioxidant and antibacterial activities of hibiscus (Hibiscus rosa-sinensis L.) and Cassia (Senna bicapsularis L.) flower extracts. J King Saud Univ – Sci. 25(4):275–282.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.