Abstract
We sequenced the complete mitogenome of the invasive flatworm Parakontikia ventrolineata (Platyhelminthes, order Tricladida, family Geoplanidae). The genome is 17,210 bp long, and displays common unusual characteristics shared with Platydemus manokwari, such as its colinearity, an overlap between ND4L and ND4 genes and an unusually long cox2 genes. Both Parakontikia and Platydemus are members of the subfamily Rhynchodeminae and their close relationships are supported by the maximum likelihood phylogeny inferred from the protein-coding genes.
Parakontikia ventrolineata (Dendy Citation1892) Winsor, Citation1991 is an invasive flatworm originating from Australia (Dendy Citation1892; Winsor Citation1991). The species is sometimes designated under the binomial Kontikia ventrolineata (Dendy Citation1892). It has been recorded in various parts of the world including the United States, Mexico, United Kingdom, Ireland, Spain and France (Alvarez-Presas et al. Citation2014; Sluys Citation2016). With its size comprised between 1 and 5 cm, it is small enough to enter inside damaged fruits, especially strawberries and apples, and as such is considered a nuisance by gardeners (Justine et al. Citation2014). Parakontikia ventrolineata has a generalist diet, including snails, slugs, and woodlice, and is also a scavenger on dead, crushed earthworms and snails (Winsor et al. Citation2004; Justine et al. Citation2014).
We sequenced the complete mitogenome of P. ventrolineata. The specimen used is part of a series of 10 specimens collected in Caen, France, on 26 March 2015 and deposited in the collections of the Muséum national d’Histoire naturelle, Paris, under number MNHN JL240 and where the rest of the specimen is kept. Paired end sequencing was conducted by the Beijing Genomics Institute (Shenzhen) on a DNBSEQ platform, resulting in a total of 60 million 100-bp reads. Assembly was performed using with SPAdes 3.14.0 (Bankevich et al. Citation2012) with a k-mer of 85. Genes were identified with the help of MITOS (Bernt et al. Citation2013).
The genome is 17210 bp long (GenBank accession number: MT081960) and it codes for 13 proteins, 2 rRNAs, and 21 tRNAs. It appears to be very similar to the recently sequenced mitogenome of Platydemus manokwari (GenBank: MT081580; Gastineau et al. Citation2020), an invasive flatworm (see Justine et al. Citation2014; Citation2015) also belonging to the subfamily Rhynchodeminae. Genomes are colinear including for the positions of tRNA, the open reading frames corresponding to the ND4L and ND4 genes are overlapping by an identical lenght of 32 nucleotides and especially the cox2 gene displays a similar overlength. The total length of the cox2 gene is 1302 bp, close to the size observed in P. manokwari (1359 bp) and twice for example the 678 bp of the invasive flatworm Bipalium kewense’s cox2 gene (GenBank: MK455837; Gastineau et al. Citation2019). This extension occurs between the same conserved protein domains as in P. manokwari.
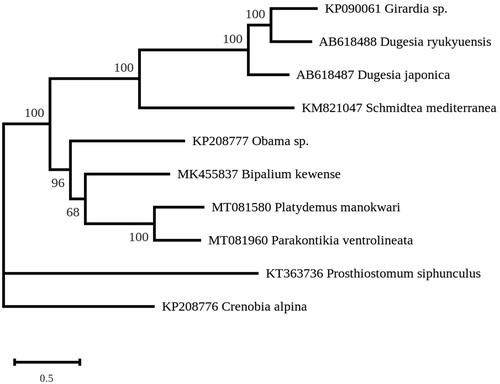
A maximum likelihood phylogeny was inferred from concatenated gene-coding proteins with the protocol and dataset of Gastineau et al. (Citation2020), using RAxML v.8.2.3 (Stamatakis Citation2014) and the MtArt substitution model (Abascal et al. Citation2007). The phylogeny strongly associates P. ventrolineata and P. manokwari in a single clade () and strictly discriminates them from the other Geoplanidae, B. kewense and Obama nungara, which are also invasive species (e.g. Justine et al. Citation2018, Citation2020). Parakontikia and Platydemus are both members of the subfamily Rhynchodeminae within the Geoplanidae. They were classified, however, in two different tribes within the Rhynchodeminae, i.e. the Rhynchodemini for Platydemus and the Caenoplanini for Parakontikia (Sluys et al. Citation2009). The family Geoplanidae includes four subfamilies, Microplaninae, Bipaliinae, Rhynchodeminae and Geoplaninae (Sluys et al. Citation2009); complete mitogenomes are now available for three of these, excluding the first one. The present findings of the shared characteristics of the cox2, ND4L and ND4 genes suggest that these are common characteristics of members of the Rhynchodeminae, separating them from the other subfamilies. Our study emphasizes the interest of complete mitogenome for the understanding of phylogeny in the Geoplanidae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors confirm that the data supporting the findings of this study are available at the following link: https://doi.org/10.5281/zenodo.3782525. The genome has also been deposited on GenBank with accession number MT081960.
Additional information
Funding
References
- Abascal F, Posada D, Zardoya R. 2007. MtArt: a new model of amino acid replacement for Arthropoda. Mol Biol Evol. 24(1):1–5.
- Alvarez-Presas M, Mateos E, Tudó A, Jones H, Riutort M. 2014. Diversity of introduced terrestrial flatworms in the Iberian Peninsula: a cautionary tale. PeerJ. 2:e430.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Dendy A. 1892. Short descriptions of new Land Planarians. Proc Roy Soc Victoria. 4: p. 35–38.
- Gastineau R, Jean-Lou Justine J-L, Lemieux C, Turmel M, Witkowski A. 2019. Complete mitogenome of the giant invasive hammerhead flatworm Bipalium kewense. Mitochondrial DNA Part B. 4(1):1343–1344.
- Gastineau R, Lemieux C, Turmel M, Justine J-L. 2020. Complete mitogenome of the invasive land flatworm Platydemus manokwari. Mitochondrial DNA Part B. 5(2):1689–1690.
- Justine J-L, Winsor L, Barrière P, Fanai C, Gey D, Han AWK, La Quay-Velázquez G, Lee BPY, Lefevre J, Meyer J, et al. 2015. The invasive land planarian Platydemus manokwari (Platyhelminthes, Geoplanidae): records from six new localities, including the first in the USA. PeerJ. 3:e1037.
- Justine J-L, Winsor L, Gey D, Gros P, Thévenot J. 2018. Giant worms chez moi! Hammerhead flatworms (Platyhelminthes, Geoplanidae, Bipalium spp., Diversibipalium spp.) in metropolitan France and overseas French territories. PeerJ. 6:e4672.
- Justine J-L, Winsor L, Gey D, Gros P, Thévenot J. 2020. Obama chez moi! The invasion of metropolitan France by the land planarian Obama nungara (Platyhelminthes, Geoplanidae). PeerJ. 8:e8385.
- Justine JL, Winsor L, Gey D, Gros P, Thévenot J. 2014. The invasive New Guinea flatworm Platydemus manokwari in France, the first record for Europe: time for action is now. PeerJ. 2:e297.
- Justine J-L, Thévenot J, Winsor L. 2014. Les sept plathelminthes invasifs introduits en France. Phytoma. 674:28–32.
- Sluys R. 2016. Invasion of the flatworms. Am Sci. 104(5):288–295.
- Sluys R, Kawakatsu M, Riutort M, Baguña J. 2009. A new higher classification of planarian flatworms (Platyhelminthes, Tricladida). J Nat Hist. 43(29–30):1763–1777.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Winsor L. 1991. A provisional classification of Australian terrestrial geoplanid flatworms (Tricladida: Terricola: Geoplanidae). Victorian Naturalist (Blackburn). 108:42–49.
- Winsor L, Johns PM, Barker GM. 2004. Terrestrial planarians (Platyhelminthes: Tricladida: Terricola) predaceous on terrestrial gastropods. In: Barker GM, ed. Natural enemies of terrestrial molluscs. Oxfordshire, UK: CAB International, p. 227–278.