Abstract
Cavia aperea is a wild guinea pig found throughout South America. The previously published mitochondrial sequence for C. aperea was highly divergent from the C. porcellus sequence and contained stop codons within open reading frames. Here we resequenced the mitochondrial genomes of C. aperea and C. porcellus. Both sequences reflect gene organization typical for mammalian mitochondrial DNA. Our C. aperea mtDNA sequence shows that all of the open reading frames are intact, but confirms the strikingly low level of sequence identity (92.7%) with the closely related C. porcellus mtDNA.
Keywords:
Cavia aperea is a wild guinea pig found through large parts of South America (Rood Citation1972). Allometric studies confirm that the laboratory strain Cavia porcellus descended from C. aperea (Kruska and Steffen Citation2013). Both C. aperea and C. porcellus have been used in behavioral research and they may provide a superior model for reproductive research to mouse because of similarities between progesterone levels in pregnancy in humans and in cavies and the potential for preterm delivery (Mitchell and Taggart Citation2009). Hybrids between aperea and porcellus have been generated for studies of chromosome structure (George et al. Citation2009), reproduction (Rood and Weir Citation1970) and enzymology (Carter et al. Citation1972). The hybrids are both viable and fertile.
The mitochondrial sequence for C. aperea was previously published (Cao and Xia Citation2015) based upon reads from a whole genome sequencing study (Weyrich et al. Citation2014). The sequence (KT439327) was highly divergent from the sequence of porcellus mtDNA. This was surprising, because mitochondrial genetic incompatibility can prevent hybridization and lead to male sterility, colorfully known as ‘mother’s curse’ (Wolff et al. Citation2014). However the published aperea sequence KT439327 also contained nonsense codons in open reading frames suggesting the possibility of error. Here we re-sequenced the mtDNA of C. aperea and C. porcellus.
C. aperea were collected in Uruguay and were housed at the University of Bielefeld Department Of Animal Behavior. C. porcellus were obtained from Charles River. DNA was amplified using primers (available upon request) based upon the published sequences. Amplified sequences were confirmed by gel electrophoresis, sequenced and assembled (Geneious Prime, Biomatters Ltd.).
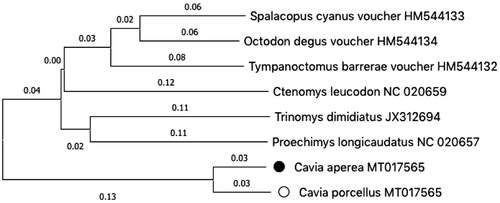
The mtDNA sequence of Cavia aperea (MT017566) and Cavia porcellus (MT017565) have been deposited in GenBank. The sequenced DNA is stored at the SickKids Central Biobank (Canadian Tissue Repository Network #BRC-00181) as S01-565 (porcellus) and S01-566 (aperea). We constructed a neighbour-joining phylogenetic tree of these sequences (). MT017565 is highly similar to the published sequence NC_000884.1 with no differences in amino acid sequence and only six non-coding substitutions in total. Our aperea mtDNA is 16,681 nucleotides in length. The sequence confirms that spurious insertions were present in KT439327. The overall gene structure of the C. aperea mtDNA is unremarkable, with no change in the canonical ordering or orientation of the coding genes. Like the previously published sequence of aperea, we found a low level of sequence identity to porcellus (92.7%). Amino acid differences were found in 12 out of the 13 protein coding genes including the following, ND1 (3), ND2 (14), ND3 (8), ND4 (15), ND5 (37), ND6 (9), ATP6 (9), ATP8 (7), COX1 (5), COX2 (4), COX3 (4), and CYTB (10) with the number in parentheses indicating the number of residue changes.
Figure 1. Phylogenetic relationship of the Cavia aperea ● and Cavia porcellus ^, and six other rodent species. Tree was constructed using the ClustalW alignment and neighbour-joining analysis in Mega X (Kumar et al. Citation2018).
This level of sequence divergence is striking. For example, the identity between the human reference sequence (rCRS) and the sequence of Neandertal mtDNA is 98.7% (Green et al. Citation2008) and the maximal known divergence between two modern human mtDNAs is 99.4% (Ingman et al. Citation2000). The low level of identity between cavia and aperea mtDNA is remarkable given their capacity to interbreed. The ability to survive the high level of incompatibility between mitochondrial and nuclear genomes requires further investigation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available on GenBank using the accession numbers MT017566 (https://www.ncbi.nlm.nih.gov/nuccore/MT017566) and MT017565 (https://www.ncbi.nlm.nih.gov/nuccore/MT017565).
References
- Cao W, Xia Y. 2015. The complete mitochondrial genome of the Cavia aperea. Mitochondrial DNA. 1394:1–2.
- Carter ND, Hill MR, Weir BJ. 1972. Genetic variation of phosphoglucose isomerase in some hystricomorph rodents. Biochem Genet. 6(2):147–156.
- George W, Weir BJ, Bedford J. 2009. Chromosome studies in some members of the family Caviidae (Mammalia: Rodentia). J Zool. 168(1):81–89.
- Green RE, Malaspinas A, Krause J, Briggs AW, Johnson PLFF, Uhler C, Meyer M, Good JM, Maricic T, Stenzel U, et al. 2008. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 134(3):416–426.
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U. 2000. Mitochondrial genome variation and the origin of modern humans. Nature. 408(6813):708–713.
- Kruska DCT, Steffen K. 2013. Comparative allometric investigations on the skulls of wild cavies (Cavia aperea) versus domesticated guinea pigs (C. aperea f. porcellus) with comments on the domestication of this species. Mamm Biol. 78(3):178–186.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Mitchell BF, Taggart MJ. 2009. Are animal models relevant to key aspects of human parturition? Am J Physiol Regul Integr Comp Physiol. 297(3):R525–R545.
- Rood JP. 1972. Ecological and behavioural comparisons of three genera of Argentine cavies. Anim Behav Monogr. 5:1–83.
- Rood JP, Weir BJ. 1970. Reproduction in female wild guinea-pigs. J Reprod Fertil. 23(3):393–409.
- Weyrich A, Schüllermann T, Heeger F, Jeschek M, Mazzoni CJ, Chen W, Schumann K, Fickel J. 2014. Whole genome sequencing and methylome analysis of the wild guinea pig. BMC Genomics. 15(1):1036.
- Wolff JN, Ladoukakis ED, Enríquez JA, Dowling DK. 2014. Mitonuclear interactions: evolutionary consequences over multiple biological scales. Philos Trans R Soc Lond B Biol Sci. 369(1646):20130443.
