Abstract
Eleutherococcus senticosus is a highly valued woody herb medicinal plant belonging to the family Araliaceae, which is also the popular edible plant in China. In this paper, the chloroplast genome of E. senticosus was completed. The complete chloroplast genome of E. senticosus was 156,768 bp in length as a circle. It contained a large single-copy (LSC) region of 86,756 bp, a small single- copy (SSC) region of 18,154 bp and separated by two inverted repeat (IR) regions of 25,929 bp. The base compositions of chloroplast genome is uneven and the overall nucleotide composition is: A (30.7%), T (31.4%), C (19.3%) and G (18.6%), with a total G + C content (39.4%). It comprised 134 genes, including 89 protein-coding genes (PCGs), 37 transfer RNA (tRNA) genes and 8 ribosomal RNA (rRNA) genes. 8 PCG genes species, 7 tRNA genes species and 4 rRNA species were found duplicated in the IR regions. The phylogenetic analysis result shown that the chloroplast genome of E. senticosus is closest to Eleutherococcus sessiliflorus of the family Araliaceae in this study by the maximum-likelihood (ML) method. The complete chloroplast genome of E. senticosus can provide more genomics information for further research on the species in China.
Eleutherococcus senticosus is a highly valued woody medicinal plant belonging to the family Araliaceae, which is also the popular edible plant in China. E. senticosus is called Ci-Wu-Jia in Chinese that grows in the Russian Far East, Northeast China, Korea and Japan (Hwang et al. Citation2015). E. senticosus is popularly known as the Siberian ginseng because of its remarkable pharmacological effects, which can be used as a tonic and sedative and to treat rheumatism and diabetes (Umeyama et al. Citation1992). The main activities of E. senticosus are contains flavonoids, organic acids, phenols, triterpenoid saponins, lignans, coumarins, polysaccharides and other compounds (Wang et al. Citation2019). E. senticosus has been widely as an herb medicine used to treat neurasthenia, hypertension, immune regulation, cerebrovascular diseases and ischemic heart disease in China (Wang et al. Citation2019). Here, the complete chloroplast genome of E. senticosus was completed, which can be useful to study this species phylogenetic relationship and also can provide more genomics information for further research on the herb species in China.
The chloroplast DNA of E. senticosus was extracted from its fresh leaves which were sampled from the herb medicine market (Jilin, China, 127.30E; 42.44 N). The corresponding voucher herbarium specimen of E. senticosus was stored in Jilin Agricultural University Information Laboratory (No. JAUIL002) and the chloroplast DNA extracted using the Plant Tissues Genomic DNA Extraction Kit (TIANGEN, BJ, and CN). The chloroplast genome DNA was purified and sequenced, which was quality controlled and removed to the collected raw sequences. The chloroplast (cp) genome was assembled and annotated using the MitoZ (Meng et al. Citation2019). All the genes were analyzed using CPGAVAS (Liu et al. Citation2012) and combined with the online alignment tool Blastx and ORF Finder (NCBI). The tRNA genes were predicted using the online sites tRNA-scan (Lowe and Chan Citation2016). For the phylogenetic analysis, we selected and downloaded other 22 complete chloroplast genome sequences from the NCBI database. All the 23 chloroplast genome sequences were plants from NCBI. The annotated complete chloroplast genome sequence had been submitted to NCBI, the accession number is MK637765.2.
The complete chloroplast genome of E. senticosus sequence (MK637765.2) was 156,768 bp in length as a circle. The gene order and structure of the E. senticosus chloroplast genome is similar to the typical plant. It contained a large single-copy (LSC) region of 86,756 bp, a small single- copy (SSC) region of 18,154 bp and separated by two inverted repeat (IR) regions of 25,929 bp. The base compositions of chloroplast genome is uneven and the overall nucleotide composition is: A of 30.7%, T of 31.4%, C of 19.3% and G of 18.6% with a total G + C content of 39.4%. The chloroplast genome of E. senticosus comprised 134 genes, including 89 protein-coding genes (PCGs), 37 transfer RNA (tRNA) genes and 8 ribosomal RNA (rRNA) genes. 8 PCG genes species (rpl23, rpl2, ycf2, ycf15, ndhB, rps7, rps12 and ycf1), 7 tRNA genes species (trnHis-GAU, trnLeu-CAA, trnVal-GAC, trnIle-GAU, trnAla-UGC, trnArg-ACG and trnAsn-GUU) and 4 rRNA species (rrn16, rrn23, rrn4.5 and rrn5) were found duplicated in the IR regions, respectively.
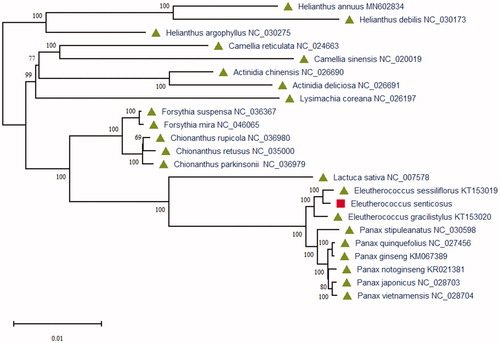
To study the phylogenetic relationships of E. senticosus, we constructed the maximum-likelihood (ML) phylogenetic tree using 23 plant species chloroplast genome sequences. Phylogenetic analysis was performed using RAxML software (Stamatakis Citation2014) with the best model, which used the number of bootstrap replicates with 2000. The ML phylogenetic tree was inferred with strong support and used the bootstrap values from 2000 replicates at all the nodes. The phylogenetic ML tree was drawn and edited using MEGA X (Kumar et al. Citation2018). As the phylogenetic analysis result (), the chloroplast genome of E. senticosus is closest to Eleutherococcus sessiliflorus (KT153019) of the family Araliaceae in the phylogenetic relationship. This chloroplast genome of E. senticosus can be used to study this species genetic diversity and phylogenetic relationship that also can provide more genomics information for further research on the herb species in China.
Data openly available in a public repository that issues datasets with DOIs
The data that support the findings of this study are openly available in Eleutherococcus senticosus at http://doi.org/[doi], reference number is MK637765.2.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available from the corresponding author and upon reasonable request.
References
- Hwang HS, Lee H, Choi Y. 2015. Transcriptomic analysis of siberian ginseng (Eleutherococcus senticosus) to discover genes involved in saponin biosynthesis. BMC Genomics. 16:180.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715–2164.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Umeyama A, Shoji N, Takei M, Endo K, Arihara S. 1992. Ciwujianosides D1 and C1: powerful inhibitors of histamine release induced by anti-immunoglobulin E from rat peritoneal mast cells. J Pharm Sci. 81(7):661–662.
- Wang RJ, Shi LQ, Liu S, Liu ZQ, Song FR, Sun ZH, Liu ZY. 2019. Mass spectrometry-based urinary metabolomics for the investigation on the mechanism of action of Eleutherococcus senticosus (Rupr. & Maxim.) Maxim. leaves against ischemic stroke in rats. J Ethnopharmacol. 241:111969.