Abstract
Lepus oiostolus is widely inhabited in the Qinghai-Tibet Plateau. So far, little mitochondrial genome information of this genus has been described. To grasp a better comprehension on the molecular basis of L. oiostolus, we obtained the complete mitochondrial DNA genome sequences of this species. The mitogenome was 17,320 bp in length, which consists of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 1 noncoding regions. The complete mitochondrial genome of L. oiostolus would be of great utility in the phylogenetic analysis of the Lagomorpha and also provide meritorious insights into the deeper problems of the phylogenic analysis.
Lepus oiostolus is an endemic species in the Qinghai-Tibet Plateau, also known as gray-tailed hare. It belongs to the genus Lepus (Hodgson Citation1840; Kao and Feng Citation1964). In this study, the sample of L. oiostolus was collected from Fenxiakou, MenYuan Country, Qinghai Province, China (37.6160 N; 101.3132E). The genome DNA was extracted from the muscle tissue, using a modified method from the standard phenol/chloroform extraction process (Sambrook et al. Citation1989). The specimen of L. oiostolus, named as HB-01 was stored in the animal specimen room in Herbarium of College of Ecological Environment and Resources, Qinghai Nationalities University.
The complete mitochondrion genome of L. oiostolus was sequenced using the next-generation sequencing with Illumina Hiseq platform. The trimmed reads were mainly assembled by SPAdes (Bankevich et al. Citation2012). The whole sequence was annotated using the software Generous v11.1.5, and tRNA genes were predicted using online software MITOS (Bernt et al. Citation2013).
The mitochondrial genome of L. oiostolus was 17,320 bp in length, of which 16,124 nucleotides are coding DNA. This mitochondrial DNA sequence has been deposited in GenBank (accession NO. MT376741). It is a double-stranded closed loop structure composed of 13 protein-coding genes (PCGs), 2 rRNAs, 22 tRNAs and a D-Loop region. The overall base composition of the whole mitochondrial genome is 32% A, 29.4% T, 25.6% C, and 13.1% G, exhibiting obvious AT bias (61.4%).
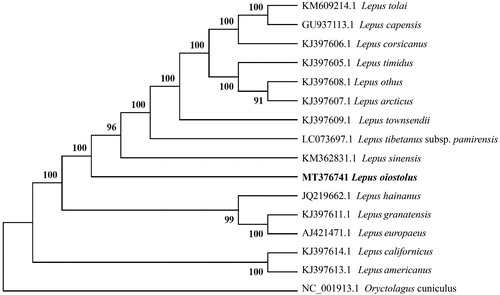
Phylogenetic relationships of L. oiostolus with 15 species of Lepus from GenBank were resolved by means of Neighbor-joining (NJ) and an Oryctolagus cuniculus as outgroup. The NJ tree was built using MEGA 7 (Kumar et al. Citation2016) with bootstrap set to 1000. The phylogenetic tree suggested that L. oiostolus, L. tolai, L. capensis, L. corsicanus, L. timidus, L. othus, L. arcticus, L. townsendii, L. tibetanus subsp. pamirensis, and L. sinensis have a closer relationship compared with other species studied (). Hares may have reticulate evolution and resulted in the rapid radiation and speciation of Lepus (Wu et al. Citation2005; Liu et al. Citation2011). This study can provide more basic data on conservation, genetics and phylogeny of L. oiostolus for the future research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in Genbank at https://www.ncbi.nlm.nih.gov/genbank/, reference number MT376741.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Hodgson BH. 1840. On the common hare of the gangetic provinces, and of the Sub-Hemalaya; with a slight notice of a strictly He,alayan species. J Asia Soc Bengal Part 1. 9:1186.
- Kao YT, Feng ZJ. 1964. On the subspecies of the chinese grey-tailed hare,lepus oiostolus hodgson. Acta Zootaxonomica Sin. 1(1):19–29.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu J, Yu L, Arnold ML, Wu CH, Wu SF, Lu X, Zhang YP. 2011. Reticulate evolution: frequent introgressive hybridization among Chinese hares (genus lepus) revealed by analyses of multiple mitochondrial and nuclear dna loci. BMC Evol Biol. 11(1):223–214.
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press.
- Wu CH, Wu JP, Bunch TD, Li QW, Wang YX, Zhang YP. 2005. Molecular phylogenetics and biogeography of Lepus in Eastern Asia based on mitochondrial DNA sequences. Mol Phylogenet Evol. 37(1):45–61.