Abstract
Betula chichibuensis is a critically endangered limestone birch confined to the Chichibu and Kitakami mountains in central and northeastern Japan, respectively. In this study, we assembled and characterized the complete chloroplast genome of B. chichibuensis. The whole chloroplast genome was 160,791 bp in length, consisting of a large single-copy (LSC) region of 89,504 bp and a small single-copy (SSC) region of 19,175 bp, separated by a pair of inverted repeat (IR) regions of 26,056 bp. It contained 133 genes, including 88 protein-coding genes (80 PCG types), 37 tRNA genes (30 tRNA types), and eight rRNA genes (four rRNA types). The overall GC content of the chloroplast genome was 36.01%. Phylogenetic analysis resolved B. chichibuensis as sister to the clade containing B. pendula.
The genus Betula L. (Betulaceae) consists of approximately 60 tree species mainly distributed in the Northern Hemisphere (Furlow Citation1990). Betula chichibuensis H. Hara (subgenus Aspera) is a small tree that grows only on limestone outcrops in the mountains (Ashburner and McAllister Citation2013). Betula chichibuensis is significantly distinct from other birches because it has ovate eglandular leaves with up to 18 pairs of lateral veins and with long hairs along the midrib and lateral veins, and clusters of numerous male catkins from several buds toward the ends of the twigs (McAllister Citation2019). This species is endemic to Japan and assessed as Critically Endangered in the IUCN Red List (Shaw et al. Citation2014). It is confined to the Chichibu and Kitakami mountains in central and northeastern Japan, respectively (Igarashi et al. Citation2017). In this study, we first report the complete chloroplast genome of B. chichibuensis, which will offer a useful resource for future conservation genetics.
Fresh leaves of B. chichibuensis were collected from a single individual along the Oku-Chichibu Forest Road (35°57´N, 138°44´E) in the Chichibu Mountains of Japan. The voucher specimen was deposited at the Herbarium of the University of Tokyo Chichibu Forest (voucher number UTCFBC00085). The total genomic DNA was extracted using the DNeasy Plant Mini Kit (QIAGEN GmbH, Hilden, Germany) and sequencing was performed on an Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA). The filtered reads were de novo assembled with NOVOPlasty (Dierckxsens et al. Citation2017). The assembled chloroplast genome was annotated using the online annotation tool GeSeq (Tillich et al. Citation2017). The annotation was checked and corrected by referring to the chloroplast genomes of related species, B. nana (KX703002) and B. costata (MN830400), using Geneious Prime 2020.0.3 (Biomatters Ltd., Auckland, New Zealand).
The complete chloroplast genome of B. chichibuensis (GenBank Accession No. LC542973) was 160,791 bp in length and had a typical quadripartite structure, consisting of a large single-copy (LSC) region of 89,504 bp and a small single-copy (SSC) region of 19,175 bp, separated by a pair of inverted repeat (IR) regions of 26,056 bp. It contained 133 genes, including 88 protein-coding genes (80 PCG types), 37 tRNA genes (30 tRNA types), and eight rRNA genes (four rRNA types). Most of the genes were located in the single-copy region, while 19 genes were duplicated in the IR regions. The overall GC content of the chloroplast genome was 36.01% while that of the LSC region, SSC region, and IRs was 33.64%, 29.50%, and 42.48%, respectively.
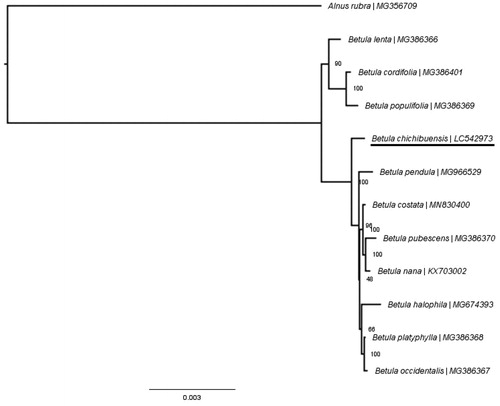
To determine the phylogenetic position of B. chichibuensis, a phylogenetic tree was constructed using the complete chloroplast genomes of B. chichibuensis, 10 Betula species and one Alnus species as the outgroup. All of the chloroplast genomes were aligned with MAFFT version 7 (Katoh et al. Citation2019), and maximum-likelihood phylogenetic inference was performed under the GTR + G + I model using RAxML-NG (Kozlov et al. Citation2019). Bootstrap values were calculated with 1000 replicates. The phylogenetic tree showed B. chichibuensis as sister to the clade containing B. pendula (). This result provides new insight for phylogenetic studies of Betula and further conservation strategies for B. chichibuensis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov/nuccore/LC542973.1, with the accession number LC542973.1.
Additional information
Funding
References
- Ashburner K, McAllister HA. 2013. The genus Betula: a taxonomic revision of birches. Kew (Richmond, UK): Royal Botanic Gardens.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Furlow J. 1990. The genera of Betulaceae in the southeastern United States. J Arnold Arbor. 71(1):1–67.
- Igarashi Y, Aihara H, Handa Y, Katsumata H, Fujii M, Nakano K, Hirao T. 2017. Development and evaluation of microsatellite markers for the critically endangered birch Betula chichibuensis (Betulaceae). Appl. Plant Sci. 5(5):1700016.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166..
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference . Bioinformatics. 35(21):4453–4455.
- McAllister H. 2019. 924. Betula chichibuensis. Curtis’s Bot Mag. 36(4):365–374.
- Shaw K, Roy S, Wilson B. 2014. Betula chichibuensis. The IUCN Red List of Threatened Species. 2014:e.T194282A2309490. https://dx.doi.org/10.2305/IUCN.UK.2014-3.RLTS.T194282A2309490.en
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.