Abstract
Bougainvillea peruviana is a widely domesticated ornamental plant species. However, studies on B. peruviana are limited. In this study, we reported the complete chloroplast (cp) genome of B. peruviana. The cp genome is 154,465 bp in length, containing a large single-copy (LSC) of 85,563 bp and a small single-copy (SSC) of 18,050 bp, which are separated by a pair of inverted repeats (IRs) of 25,426 bp, each. A total of 132 genes, including 86 protein-coding genes, 38 tRNA genes, and eight rRNA genes, were predicted. The overall GC content for the cp genome is 36.5%. The maximum-likelihood tree constructed based on cp genome sequences showed that B. peruviana is placed under Nyctaginaceae and is diverged before Bougainvillea glabra and Bougainvillea spectabilis under strong bootstrap support.
Bougainvillea is a versatile and remarkable ornamental plant widely used in the gardens (Tripathi et al. Citation2017). Among all Bougainvillea species, Bougainvillea peruviana, which is originated from Peru, is regarded as one of the major Bouganvillea species responsible for the vast applications in urban landscaping (Salam et al. Citation2017). Due to its attractive and colorful bracts, many crosses among various species have produced new hybrid species and important cultivars, wherein hybrids such as Bougainvillea × buttiana and Bougainvillea × spectoperuviana are common Bougainvillea hybrid species derived from B. peruviana (Salam et al. Citation2017). Although the species has been domesticated in many countries, information on this species is scarce (Abarca-Vargas and Petricevich Citation2018). The commercial value of B. peruviana has overshadowed genetic studies on this important horticultural species. Therefore, in this study, we characterized the complete chloroplast (cp) genome sequence of B. peruviana using the next-generation sequencing technique.
DNA extraction was carried out using the fresh leaf samples collected from the B. peruviana planted in the Germplasm Resource Nursery of Ornamental Plants, Guangzhou Institute of Forestry and Landscape Architecture (GIFLA), Guangdong Province of China (N113°20′25′′, E23°13′47′′). The voucher specimen (voucher record number: GIFLA-Bope-2019-08-30) was deposited at the voucher collection room of GIFLA. A 300-bp insert size genomic library was constructed using TruSeq DNA Sample Prep Kit (Illumina, San Diego, CA) and sequencing was conducted on an Illumina Novaseq platform. Approximately 6 GB of raw data of 150-bp paired-end reads were generated and assembled using NOVOPlasty (Dierckxsens et al. Citation2017), using the rbcL gene sequence of Bougainvillea glabra (GenBank accession number: MG833637) as the seed sequence. Genome annotation was conducted using GeSeq (Tillich et al. Citation2017) and manually corrected.
The complete cp genome sequence of B. peruviana (GenBank accession number: MT407463) is 154,465 bp in length, containing a large single-copy (LSC) region (85,563 bp), small single-copy (SSC) region (18,050 bp), and a pair of IRs (25,426 bp, each). A total of 132 genes were predicted, including 86 protein-coding genes, 38 tRNAs, and 8 rRNAs. The total GC content of the cp genome is 36.5%.
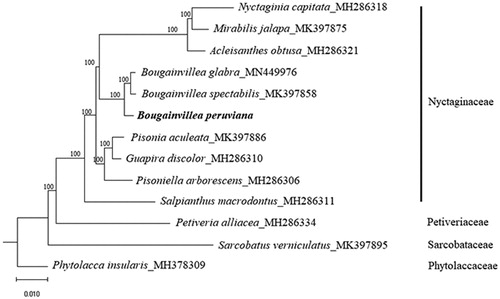
Phylogenetic analysis was conducted using RAxML available in the CIPRESS Science Gateway web portal (Miller et al., Citation2010), using 1000 bootstrap replicates, based on the cp genome sequences of 10 species in the family Nyctaginaceae, and three species, Petivera alliacea (Petiveriaceae, MH286334), Sarcobatus verniculatus (Sarcobataceae; MH376309), and Phytolacca insularis (Phytolaccaceae; MH376309), were included as an outgroup. The maximum-likelihood tree showed that B. peruviana is clustered with other members from the family Nyctaginaceae, and is diverged before Bougainvillea glabra and Bougainvillea spectabilis under strong bootstrap support (100%; ).
Figure 1. Maximum-likelihood tree based on the chloroplast genome sequences of 10 species from the family Nyctaginaceae, with Petiveria alliacea (Petiveriaceae), Sarcobatus vermiculatus (Sarcobataceae), and Phytolacca insularis (Phytolaccaceae) as outgroup. Shown next to the nodes are bootstrap support values based on 1000 replicates.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the NCBI GenBank at http://www.ncbi.nlm.nih.gov, accession number MT407463.
Additional information
Funding
References
- Abarca-Vargas R, Petricevich VL. 2018. Bougainvillea genus: a review on phytochemistry, pharmacology, and toxicology. Evid Based Complement Alternat Med. 2018:9070927.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Miller MA. Pfeiffer W. Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November, 2010. New Orleans, LA pp 1–8
- Salam P, Bhargav V, Gupta YC, Nimbolkar PK. 2017. Evolution of Bougainvillea (Bougainvillea Commers.) - a review. J Appl Nat Sci. 9(3):1489–1494.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Tripathi S, Singh S, Roy RK. 2017. Pollen morphology of Bougainvillea (Nyctaginaceae): a popular ornamental plant of tropical and sub-tropical gardens of the world. Rev Palaeobot Paly. 239:31–46.
