Abstract
Isodon lophanthoides var. graciliflorus is a folk medicinal herb that is distributed in tropical and subtropical Asia. In this study, the complete plastome of I. lophanthoides var. graciliflorus was assembled and annotated. The plastome is 152,195 bp in length, consisting of a large single-copy (LSC) region of 83,095 bp, a small single-copy (SSC) region of 17,699 bp, and two inverted repeat (IR) regions of 25,701 bp, each. It has 113 genes, including 80 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. The overall GC content of the plastome is 37.6%. Phylogenetic analysis showed that I. lophanthoides var. graciliflorus was sister to Isodon serra.
Isodon lophanthoides var. graciliflorus (Benth.) H. Hara, which is distributed in China, India, Laos, Myanmar, Nepal, Thailand and Vietnam, belongs to the genus Isodon in the family Lamiaceae (Wu and Li Citation1977; Li and Hedge Citation1994). Isodon lophanthoides var. graciliflorus, together with Isodon lophanthoides var. lophanthoides and Isodon serra (Maximowicz) Kudô, has been considered as botanical source of the traditional medicine Xihuangcao, which is commonly used in South China as a dampness-draining, anicteric and liver protection herb (Guangdong Food and Drug Administration Citation2011). However, most of the Xihuangcao-based products do not specify the taxon used. Additionally, due to the different secondary metabolites of these taxa, it is controversial whether they can be used equally (Wong et al. Citation2016). In this study, we characterized the complete plastome of I. lophanthoides var. graciliflorus to better survey the resources of Xihuangcao in the future.
The plant material of I. lophanthoides var. graciliflorus was sampled from Hutchison Whampoa Guangzhou Baiyunshan Chinese Medicine Co., Ltd (N23°11′11′′, E113°15′57′′). The voucher specimen (Zhy-Z2) was deposited at South China Botanical Garden Herbarium. Total genome DNA was extracted from fresh leaves using a modified CTAB protocol (Doyle and Doyle Citation1987). Library construction and paired-end sequencing were performed by the Beijing Genomics Institute (Wuhan, China). Genome sequences were assembled in SPAdes v3.10.1 (Bankevich et al. Citation2012); and Geneious Prime 2019 (Biomatters, Ltd., Auckland, New Zealand) was subsequently used to close gaps. The genes in the plastome were annotated and manually adjusted using Geneious Prime 2019. Phylogenomic analysis was performed using the complete plastomes from 24 taxa in the Lamiaceae (including I. lophanthoides var. graciliflorus in this study). The plastome sequences were aligned using MAFFT (Katoh and Standley Citation2013). The phylogenetic tree was constructed with RAxML (Stamatakis Citation2014), using the maximum-likelihood algorithm.
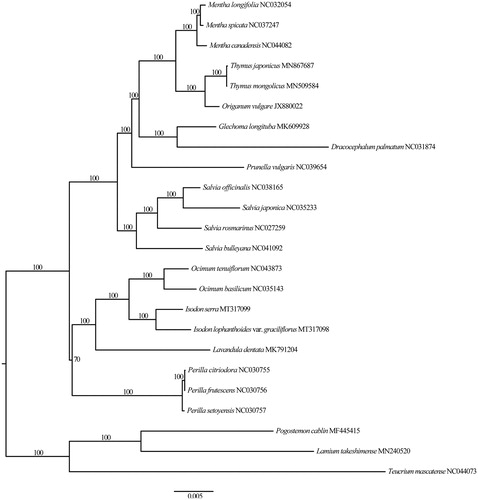
The I. lophanthoides var. graciliflorus plastome (GenBank accession no.: MT317098) is 152,195 bp in length, consisting of a large single-copy (LSC) region of 83,095 bp, a small single-copy (SSC) region of 17,699 bp, and two inverted repeat (IR) regions of 25,701 bp, each. The plastome of I. lophanthoides var. graciliflorus contains 113 genes, including 80 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. The overall GC content is 37.6%, higher than those of LSC (35.6%) and SSC (31.0%) regions, but lower than those of IR regions (43.1%). Based on the ML tree, all sampled members of tribe Ocimeae formed a clade, and I. lophanthoides var. graciliflorus was sister to I. serra (). The plastome of I. lophanthoides var. graciliflorus is of significance for its evolutionary studies and medicinal researches.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/, reference number MT317098.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Guangdong Food and Drug Administration. 2011. Guangdong Chinese Materia Medica Standards. Guangdong (China): Guangdong Science & Technology Press; p. 347–353.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li XW, Hedge IC. 1994. Lamiaceae. In Flora of China. Beijing (China): Science Press; St. Louis (MO): and Missouri Botanical Garden; p. 50–299.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wong LL, Liang ZT, Chen HB, Zhao ZZ. 2016. Rapid differentiation of Xihuangcao from the three Isodon species by UPLC-ESI-QTOF-MS/MS and chemometrics analysis. Chin Med. 11:48.
- Wu CY, Li HW. 1977. Flora Reipublicae Popularis Sinicae. Beijing (China): Science Press, 66: p. 482.